Ribosomal Ribonucleic Acid Hybridization for Organism Identification
a technology of ribosomal ribonucleic acid and hybridization, which is applied in the field of hybridization, can solve the problems of delay and the possibility of false negative, the possibility of false positive, and the particular problem of relying on slow culture methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
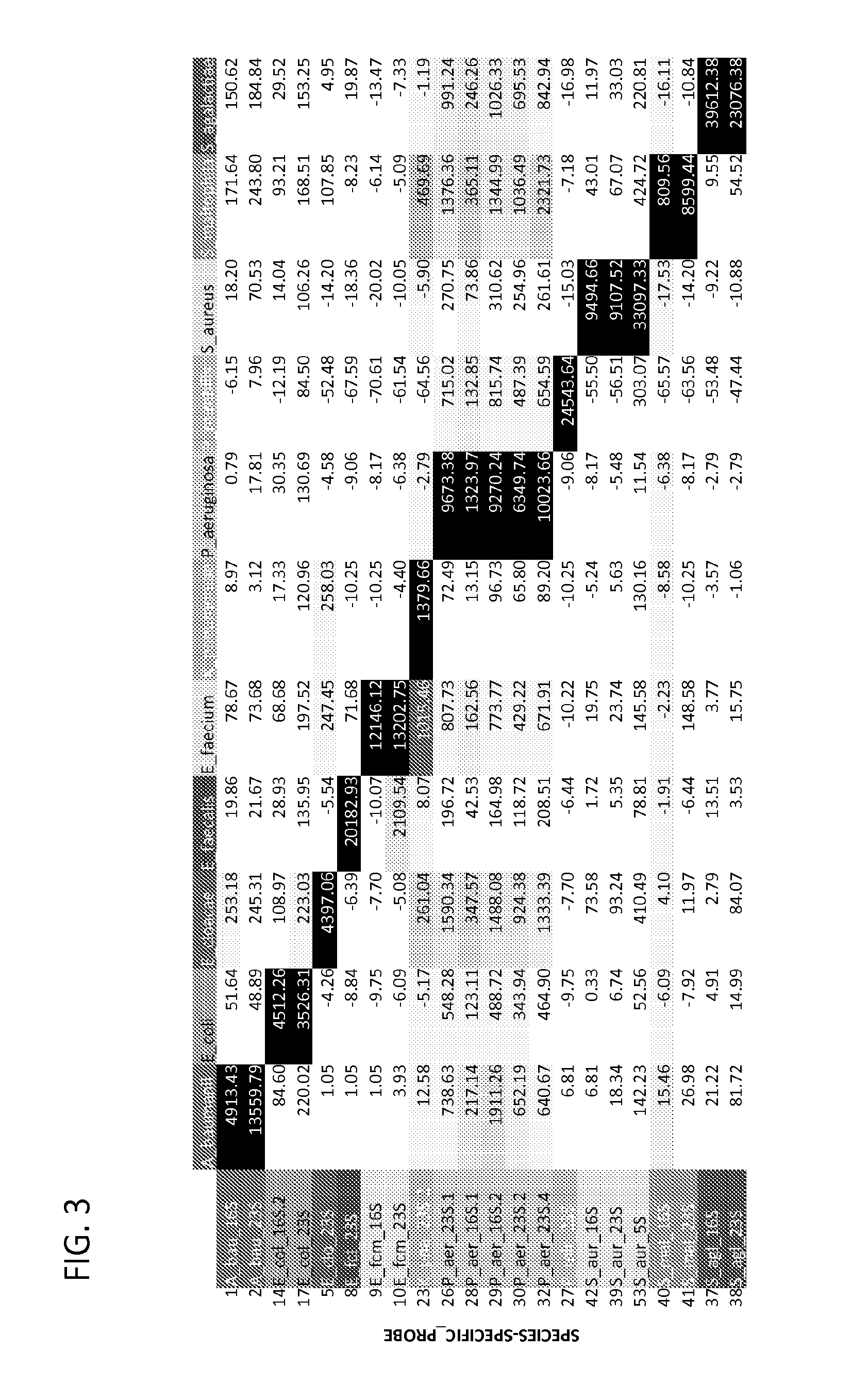
[0167]Sequence data from closely related organisms was analyzed and aligned to see if there were any stretches of 100 nucleotides with sufficient sequence divergence to allow for selective hybridization. For even the most-related organisms, Escherichia coli and Klebsiella pneumoniae, there were such regions.
[0168]Sequences were gathered from every ribosomal RNA gene (up to approximately 5-6 per genome for each subunit) from all sequenced genomes in the NCBI database for the four organisms in the pilot (Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus aureus). Of the 23S rRNA sequences, 133 sequences were derived from Escherichia coli, 23 from Klebsiella pneumoniae, nine from Pseudomonas aeruginosa, and 54 from Staphylococcus aureus; of the 16S rRNA sequences, 125 were derived from Escherichia coli, 21 from Klebsiella pneumoniae, eight from Pseudomonas aeruginosa, and 55 from Staphylococcus aureus; of the 5S rRNA sequences, 68 were de...
example 2
Trials Using the Pilot Probeset
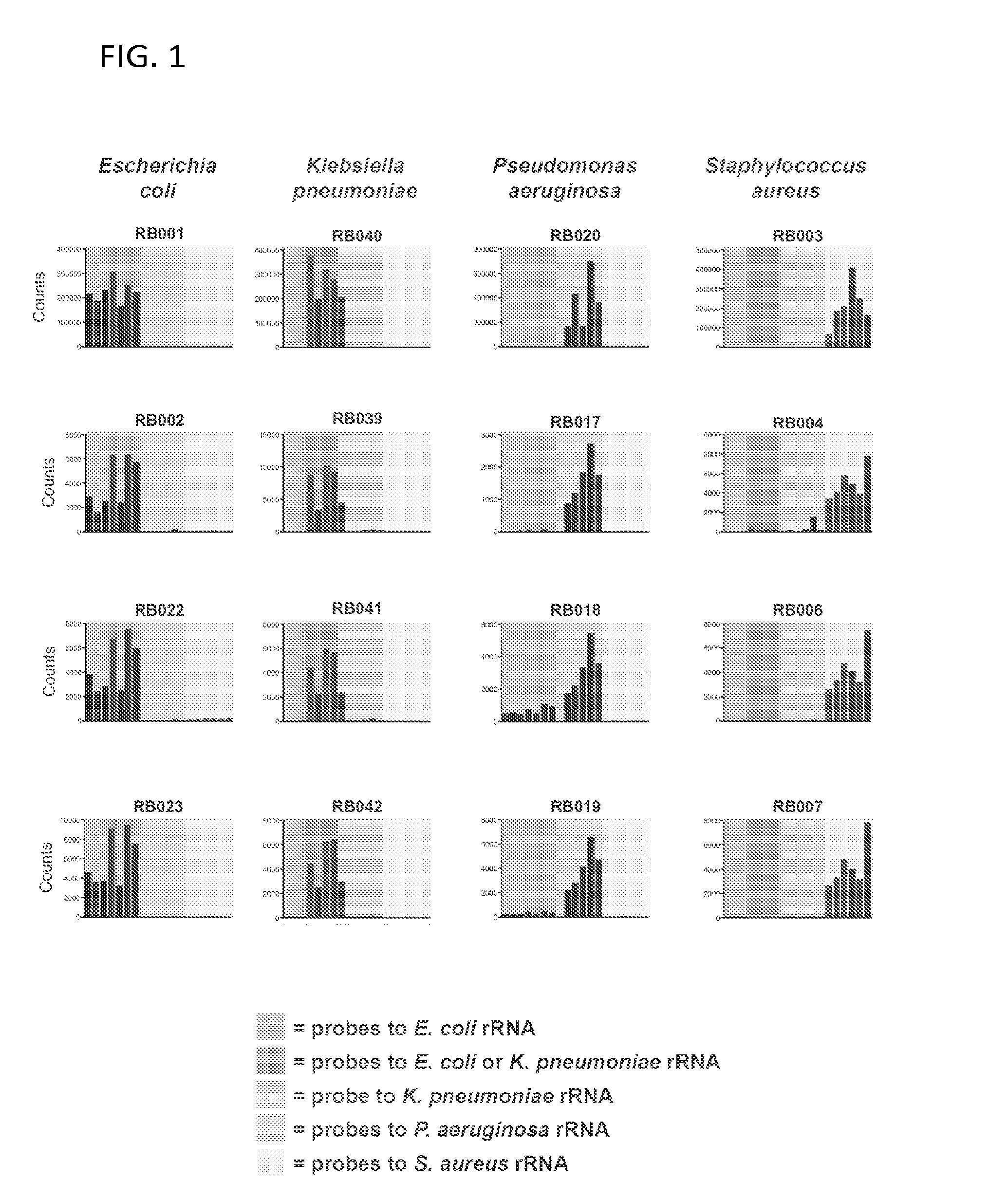
[0171]Capture and reporter probes were designed that selectively anneal to the 5S, 16S, and 23S rRNA of Escherichia coli, Klebsiella pneumoniae, Pseudomonas aeruginosa, and Staphylococcus aureus, four major clinical pathogens. The pathogens were chosen for the pilot study because they cover a broad range of phylogenetic space among pathogenic microbes: two closely related enteric Gram negative bacilli (E. coli and K. pneunzoniae) and a non-enteric Gram negative bacillus (P. aeruginosa) of the proteobacteria family, and a Gram positive coccus (S. aureus) of the firmicute family. Data from a total of 19 probes that recognize these species was generated and analyzed. The close phylogenetic relationship between the two enteric Gram negative bacilli required that 5 of these probes recognize both E. coli and K. pneumoniae, although we were also able to generate 3 probes specific to E. coli alone, and 1 probe unique to K. pneumoniae.
[0172]Referring to FIG. 1...
example 3
Dilution Series of Clinical Isolates Demonstrates Sensitivity of rRNA Recognition by Nanostring Detection
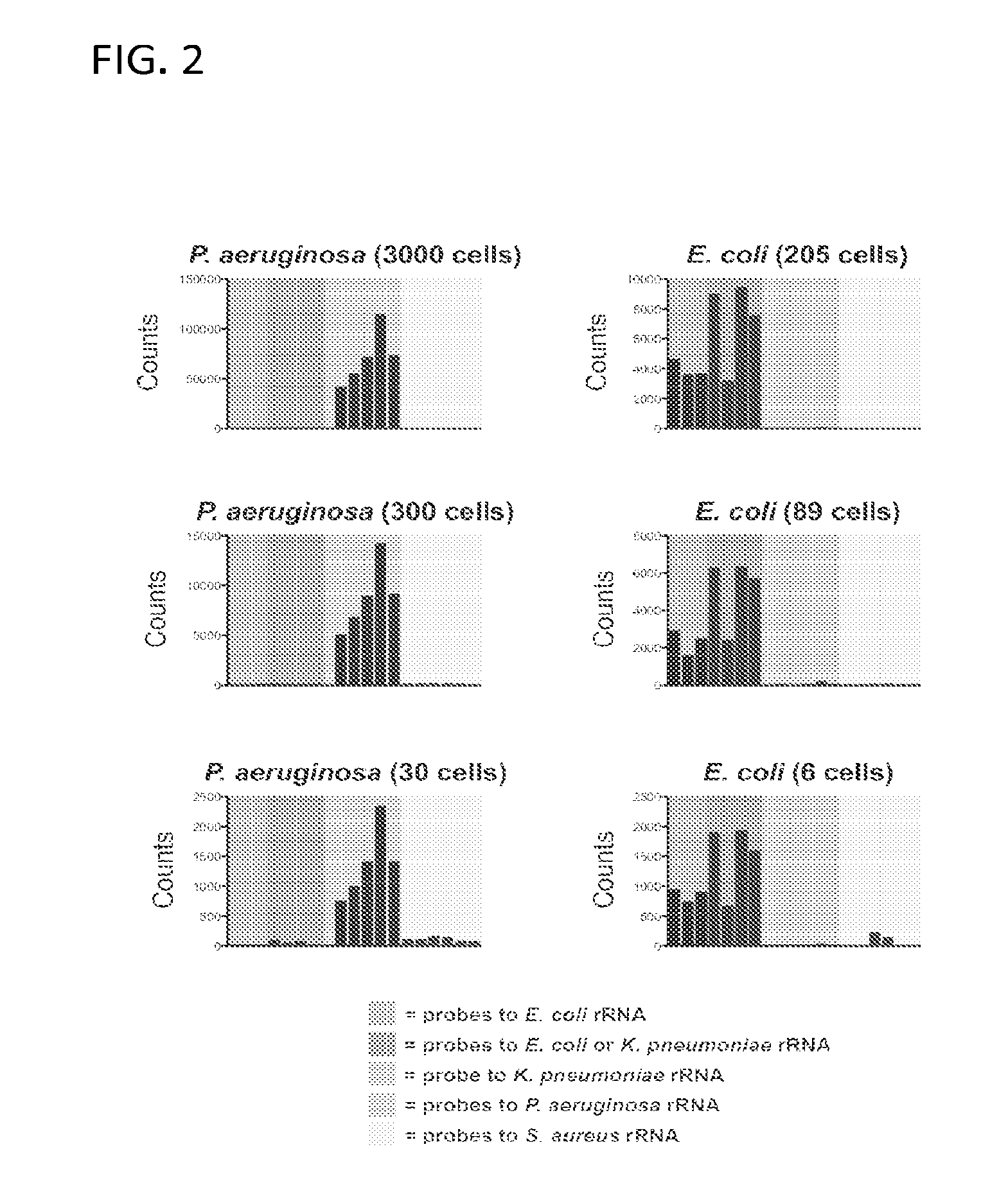
[0174]Referring now to FIG. 2, trials were carried out and results plotted as in FIG. 1, per NanoString protocol. P. aeruginosa trials (left column) were performed from axenic culture, while E. coli trials (right column) were performed from spiked blood samples purified on a spiral microchannel device. Cell count was confirmed by colony forming units.
[0175]To illustrate the power of targeting the highly abundant ribosomal RNA transcripts, samples were progressively diluted containing the bacteria of interest to test the limits of detection of this assay (FIG. 2). It was possible to detect as few as 30 Pseudomonas bacilli from axenic culture (left panels) with excellent specificity. In separate experiments using spiked blood samples from which the bacteria were separated using a spiral microfluidic device, the limits of detection was pushed even further, specifically recognizing a...
PUM
| Property | Measurement | Unit |
|---|---|---|
| time | aaaaa | aaaaa |
| absorbance wavelength | aaaaa | aaaaa |
| absorbance wavelength | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com