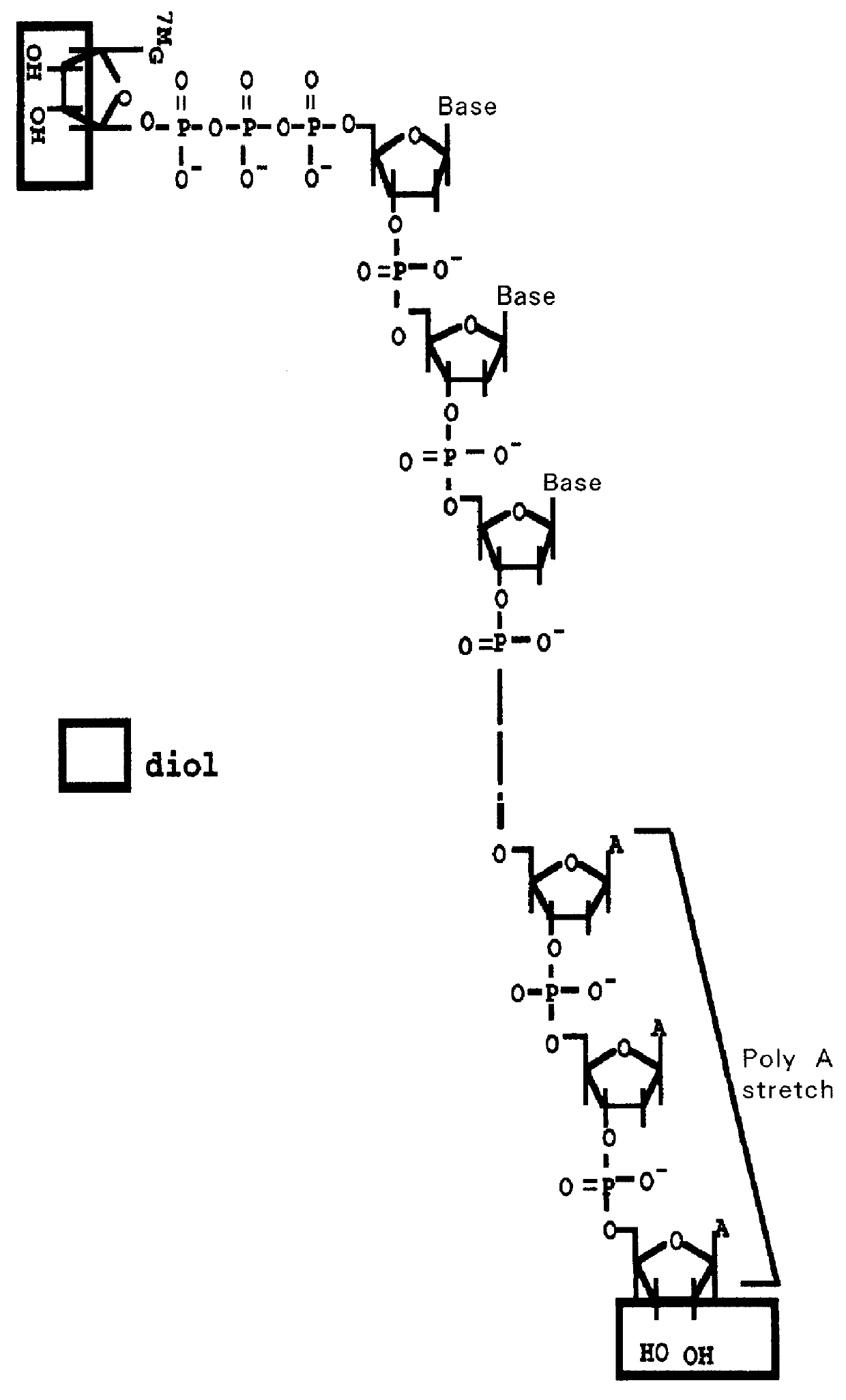
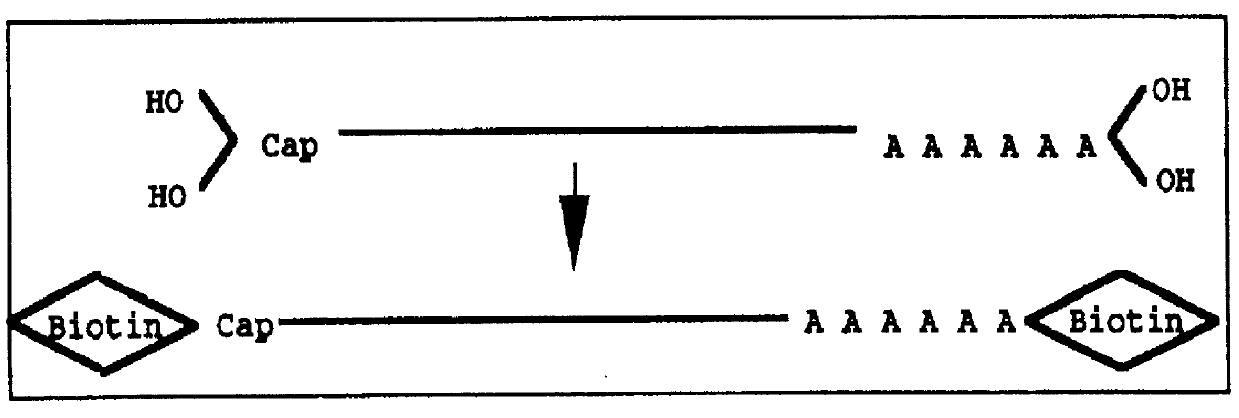
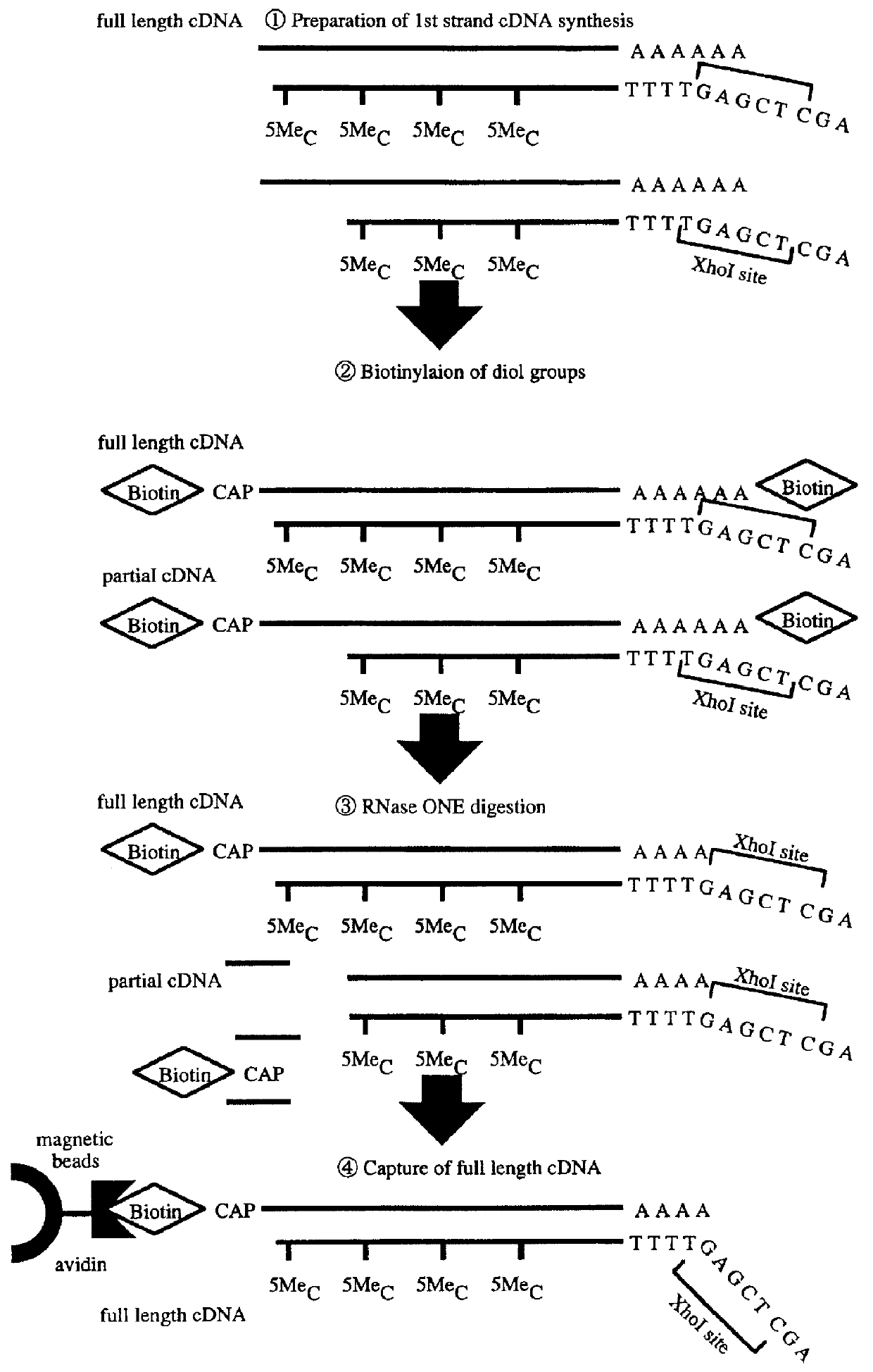
Method for forming full-length cDNA libraries
a cdna library and full-length technology, applied in the field of medical science and biology, can solve the problems of inability to analyze the whole structure of transcription units, the inability to isolate the site, and the efficiency of any conventional method for isolating cdnas in their full-lengths,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
reference example 1
With a library of double stranded full-length cDNAs obtained by the method of Japanese Patent Application No. Hei 8-60459 / 1998 (the previous method) using mRNA obtained by the same procedure as those of Example 1, lengths of inserted cDNAs were measured and tallied. The results are shown in Table 2 above. Further, double stranded full-length cDNAs were prepared by a method described below.
Synthesis of mRNA-cDNA hybrids
Reverse transcription reaction was performed by using 10 .mu.l of mRNA and 2000 units of Superscript II (Gibco BRL) in 100 .mu.l of a buffer (50 mM Tris-HCl, 75 mM KCl, 3 mM MgCl.sub.2, 10 mM DTT) in the presence of 0.5 mM 5-methyl-dCTP, 1 mM dATP, 1 mM dTTP and 1 mM dGTP. 5 .mu.g of oligonucleotide 3'NMTTTTTTTTTTTTGAGCTCTGAATCAAGAGAGAGAGAGAGAGAGAG5' (N: arbitrary nucleotide, M: G, A or C) was used as a primer. The reaction was performed at 42.degree. C. for 45 minutes, then the reaction solution was incubated at 50.degree. C. for 20 minutes to synthesize mRNA-cDNA hyb...
example 2
In the present Example, in order to demonstrate that chemical cleavage of mRNA occurring during aldehydration of the diol structure necessary for labeling mRNAs with tag molecules can be avoided by RNA-DNA hybrid structure, and as a result a synthesis efficiency of full-length cDNAs can be increased, results of autoradiograpy after denatured agarose gel electrophoresis of RNA-DNA hybrid obtained in 4 different kinds of processes as shown lanes 1-4 are shown in FIG. 5 (size markers are .lambda. Hind III).
In the results shown in FIG. 5, comparing lane 1 to lane 2, long chain products are observed more in lane 1 than those in lane 2 are. It is understood that chemical cleavage of mRNA is inhibited by RNA-DNA hybrid structure formed by cDNA synthesis made in advance. Further, comparing lane 3 to lane 4, long chain products are observed more in lane 3 than those in lane 4 are. It is understood that chemical cleavage of mRNA is inhibited by RNA-DNA hybrid structure formed by cDNA synthesi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Digital information | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
| Temperature | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com