Single protein production in living cells facilitated by a messenger RNA interferase
A cell protein and living cell technology, applied in the field of endoribonuclease, can solve the problems of cell death, severe reduction of protein synthesis, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
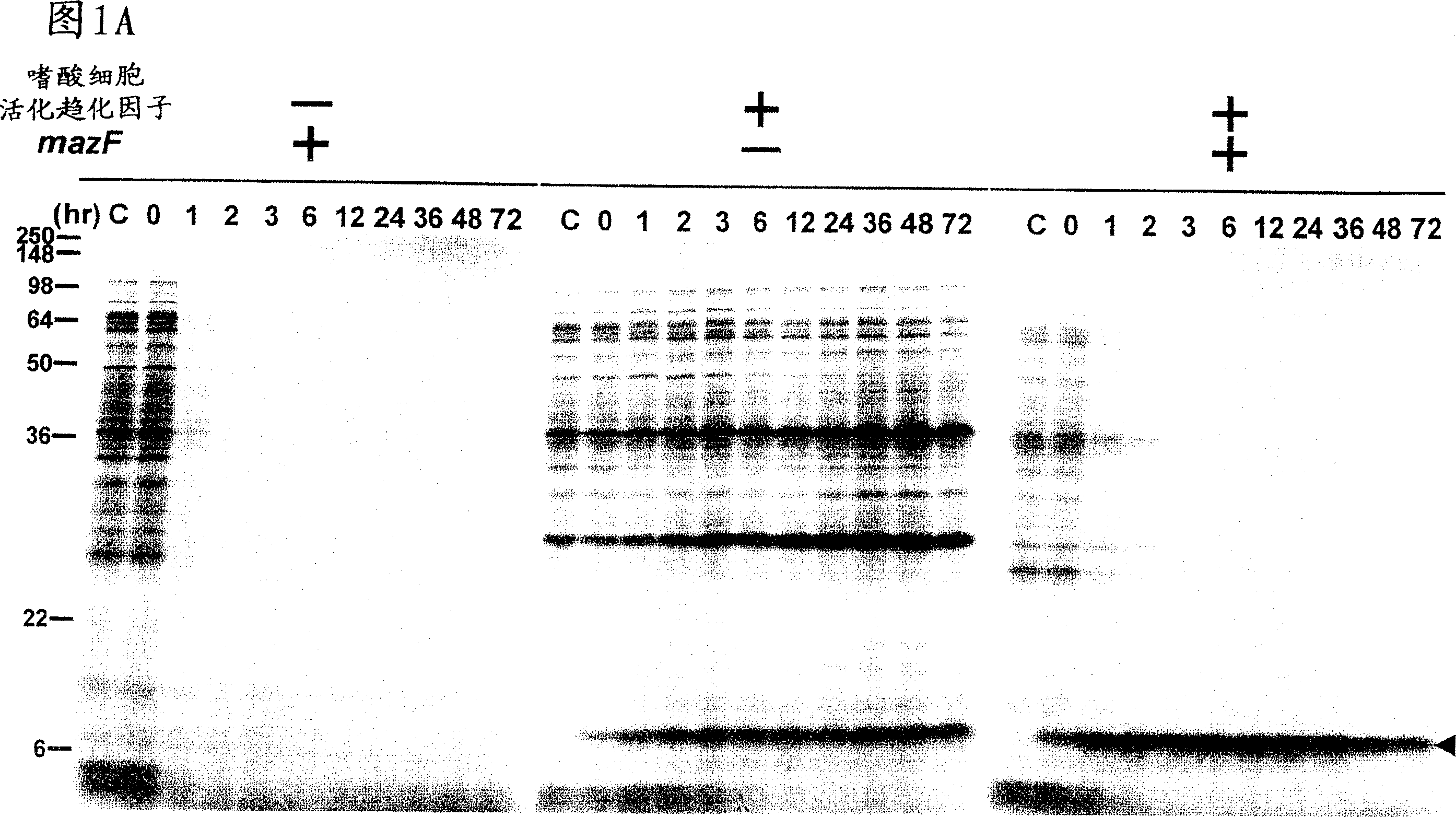
[0056] Example 1: Effect of MazF on Inducing Cellular Protein Synthesis
[0057] Large intestine harboring pACYCmazF was transformed with pColdI(SP-1) eotaxin (left panel of A and B) or pColdI(SP-2) eotaxin (right panel of B and C) Bacillus BL21 (DE3). Cells were grown in M9 glucose medium at 37°C. When OD 600 When 0.5 was reached, the culture was transferred to 15°C, and after acclimatization of the cells to the low temperature by incubation at 15°C for 45 minutes, IPTG (1 mM) was added to induce eotaxin and MazF expression (0 time). At the time points indicated above each gel, use 35 Cells were pulse-labeled with S-methionine for 15 minutes, and total cellular protein was analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) followed by autoradiography.
[0058] The mazF gene was cloned into pACYC (a low copy number plasmid containing the IPTG-inducible phage T7 promoter) to generate pACYCmazF. Cloning techniques can generally be found in J. Sambrook and D.W. Russel...
Embodiment 2
[0061] Example 2: Expression of ACA-free mRNA in MazF-induced cells
[0062] We speculate that if an mRNA engineered not to contain the ACA sequence is expressed in MazF-induced cells, this mRNA may be stably maintained in the cell such that the protein encoded by the mRNA can be produced without any other cellular protein. To test this possibility, we synthesized the gene for human eotaxin by eliminating all ACA sequences within the gene without altering the amino acid sequence. Figure 2A shows the amino acid sequence of human eotaxin and the nucleotide sequence of its gene. In the following experiments, nucleotide sequences were designed using preferred E. coli codons, and those triplets underlined were changed to ACA. Among the 64 possible triplet sequences, the ACA sequence is unique in that it can be altered to other MazF non-cleavable sequences without changing the amino acid sequence of the protein, regardless of the in-frame position of the ACA sequence.
[0063] The...
Embodiment 3
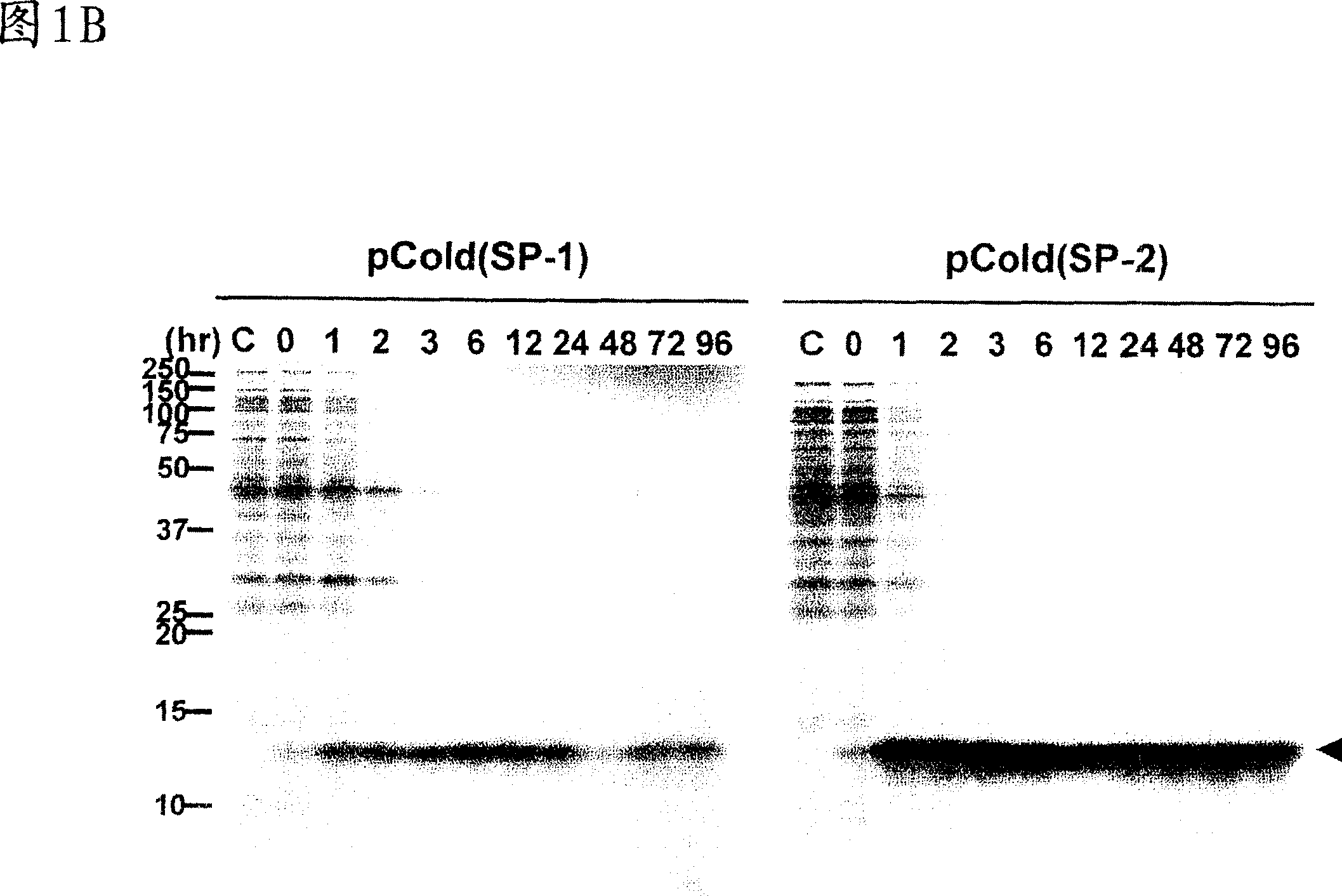
[0069] Example 3: Negative effect of ACA sequence on protein production
[0070] To verify that MazF-induced intracellular eotaxin production observed in Figure 1 was due to the ACA-free mRNA of eotaxin, into the eotaxin gene Five natural ACA sequences were added without changing their amino acid sequence as shown in Figure 2A. Utilize pColdI(SP-2) to express the eotaxin gene, and process and use [ 35 S]-methionine labeled cells. The left panel shows the results for the eotaxin gene without ACA (same as the left panel of Figure IB) and the right panel shows the results for the eotaxin gene with 5 ACA sequences.
[0071] When the gene was expressed together with pColdI(SP-1) and pACYCmazF under the same conditions as described in Figure 1, only the first 2 hours were observed compared to mRNA expression without ACA (Figure 2B, left panel). Low levels of eotaxin production were reached, after which levels further decreased to background levels (Fig. 2B, right panel).
[0072...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com