Primers, probes and methods for nucleic acid amplification
A nucleic acid and probe technology, applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 11
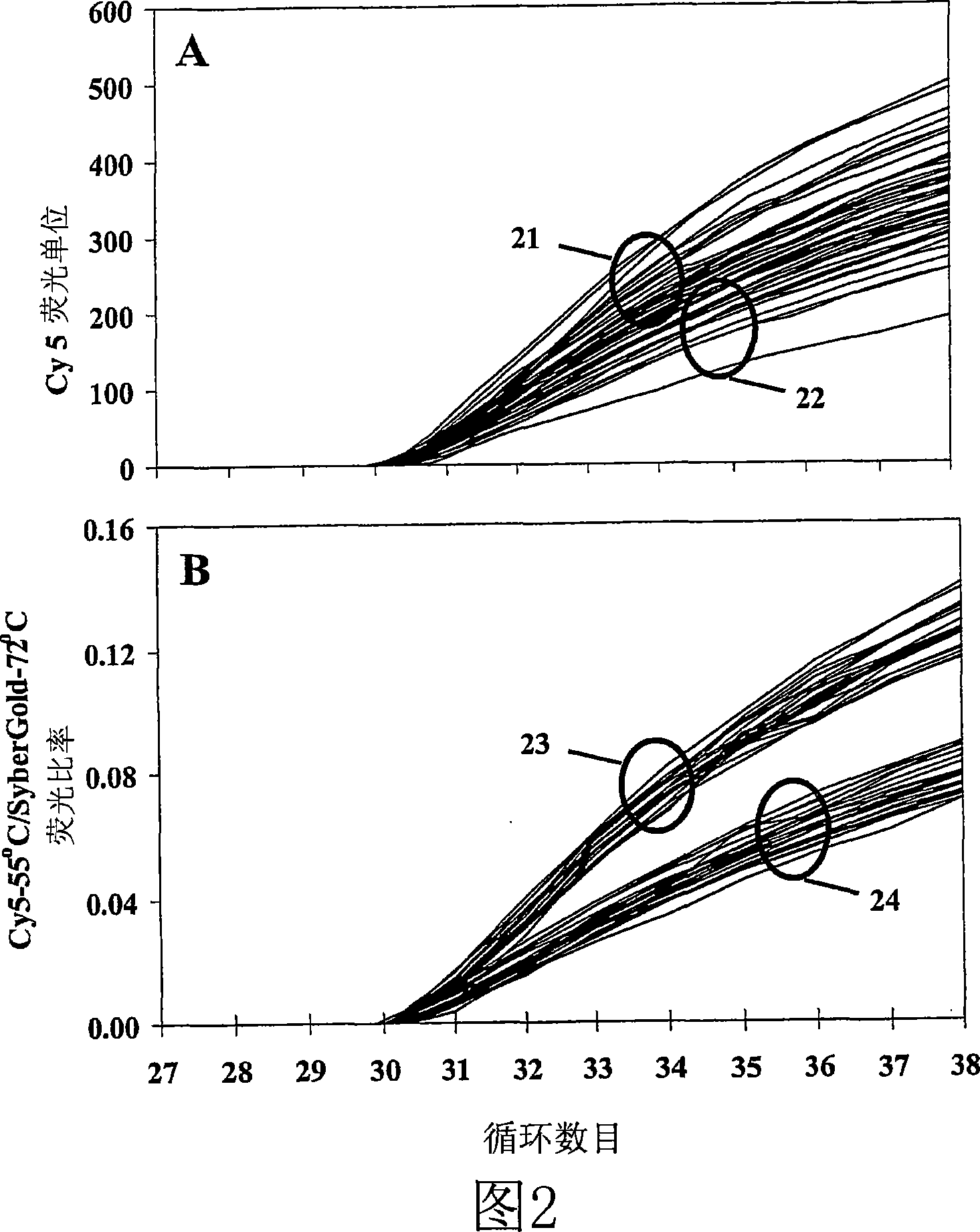
[0112] Example 11 and Figures 18-19 illustrate strategies for LATE-PCR amplification of more than one product from the same DNA template in the same reaction. Thus, these reactions contain two pairs of primers (each pair consisting of an excess primer and a limiting primer) that amplify two separate sequences within a contiguous template. The two pairs of primers can be configured such that both the excess primer and both the limiting primer hybridize to the same strand of the template, or to opposite strands of the template. As will be appreciated by those skilled in the art, two excess primers may extend "in" or "out" on their respective template strand when similar primers hybridize to opposite strands of the template. Figure 19 also shows the sequences of both excess primer strands that can be obtained from the same reaction mixture via the "dilution and removal" method.
[0113] Example 12 and Figure 20 demonstrate that the amounts of ssDNA and dsDNA produced by LATE-PCR...
example 14
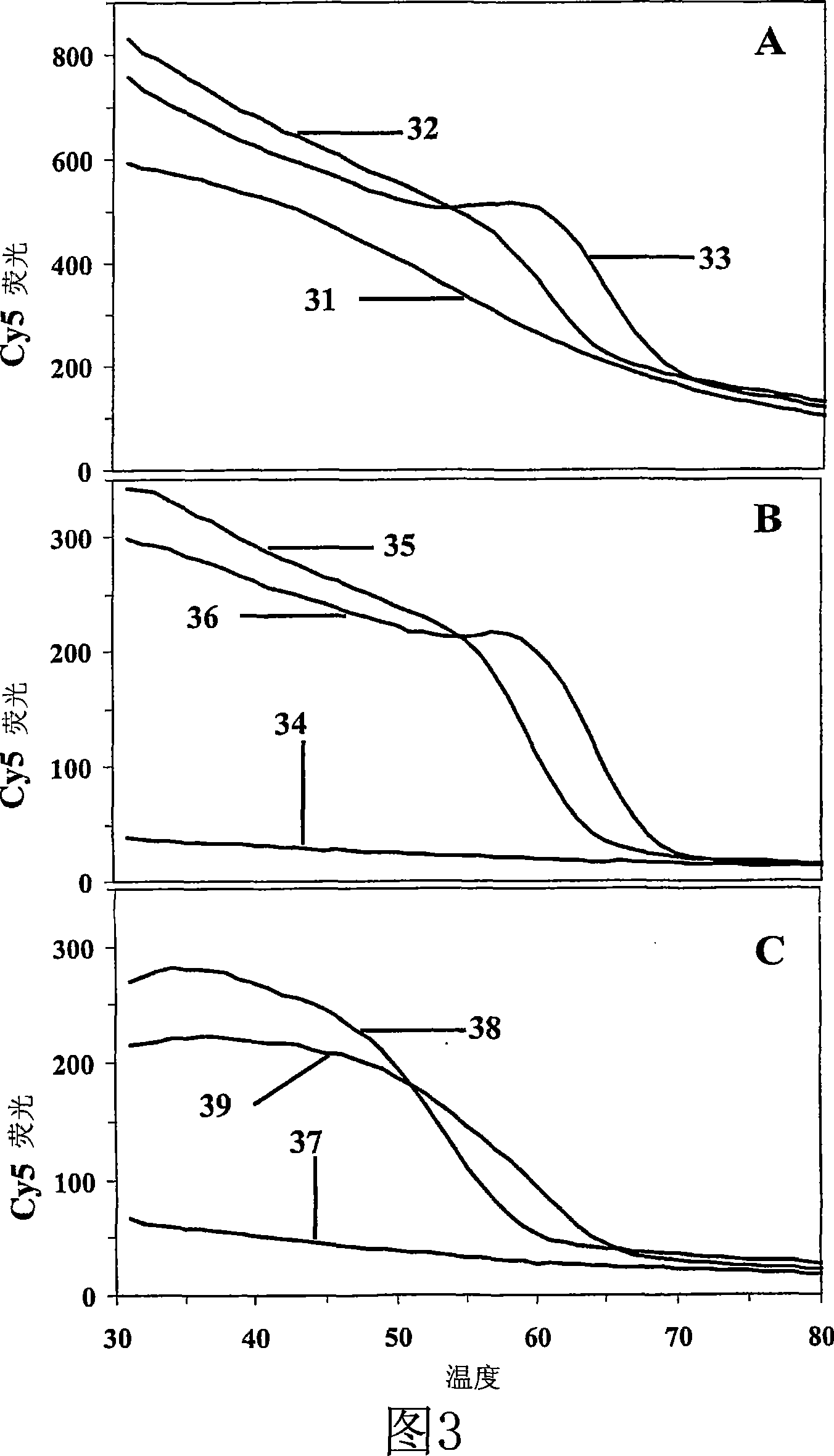
[0115] Example 14 and Figure 23 demonstrate that LATE-PCR together with at least one single mismatch-tolerant probe can be used to generate endpoint melting curves which in turn can be used to quantify the mixing of two or more closely related but distinct sequences. Relative amounts of LATE-PCR amplicons. Quantitative end-point dissolution analysis (QE) of LATE-PCR for mixtures of related amplicons is made possible by the fact that LATE-PCR produces single-stranded products. Thus, when one or more labeled mismatch-tolerant probes are present in a reaction, the probes hybridize first to the most complementary target sequence, and then if the temperature is sufficiently lowered, to all related target sequences. Each probe / target hybrid in the set thus has its own melting temperature, and the magnitude of the melting peak derived from each probe / target hybrid accurately reflects the amount of each accumulated target sequence. Quantitative measurements of the amplitude or two-di...
example 1
[0118] Example 1. Conjugated Dye vs. Conjugated Dye Plus Labeled Primer
[0119] To compare the performance of the incorporated dye with that of the dye used in conjunction with the primer comprising the interacting fluorophore, an extension assay was performed. The dye utilized was SYBR Green I at a dilution of 1:40,000.
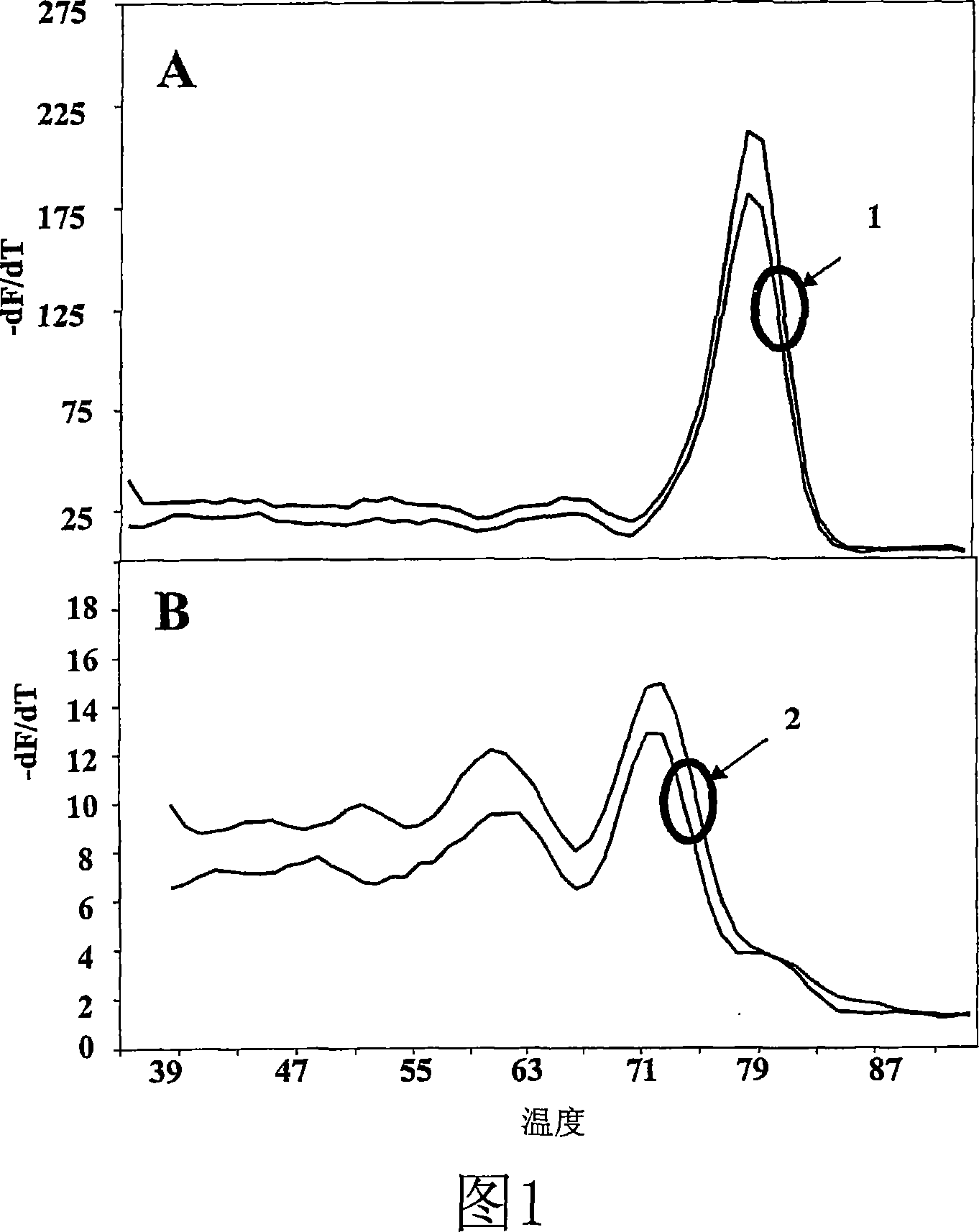
[0120] Contains three nucleotide chains. DNA template, extendable DNA primer (5' labeled with Cy5, complementary to the template, and having a T of 60°C m ), and a non-extensible DNA oligonucleotide (blocked with a phosphate group at the 3′ end), which is also complementary to the target, complementary to the primer at position 3′, also labeled with a Cy5 fluorophore, and has a relatively high temperature of 79°C High T m . The spacing between the primer and the non-extendable nucleotide is chosen such that the products of primer extension determined by the non-extendable oligonucleotide will all have a T below 79°C m .
[0121] The reaction mixture f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com