Method for detecting expression of rice blast relevant gene in infected rice leaf tissue
A disease-related gene and rice blast fungus technology, applied in biochemical equipment and methods, microbial determination/inspection, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] (1) culturing Magnaporthe grisea in liquid and extracting the total RNA of its mycelia, and reverse-transcribing the mRNA therein into cDNA;
[0021] (2) Inoculate the spores of Magnaporthe grisea on the leaves of rice susceptible variety Lijiang Xintuan Heigu, and extract the total RNA of the infected tissue at different time periods early after inoculation and reverse transcribe the mRNA therein into cDNA;
[0022] (3) Synthesis of cDNA: Take 2.5 μg of each total RNA sample, add 0.5 μg / μl Oligo (dT) 2.5 μl, 10 mM dNTP 2.5 μl, RNase-free water to 30 μl, 65 ° C water bath for 5 minutes, and quickly place in Cool on ice, add 5×First-Strand Buffer 10μl, 0.1M DTT (dithiothreitol) 5μl, 40U / μl RNase inhibitor 2.5μl, 42℃ water bath for 2μmin, then add SuperScript II reverse transcriptase 2.5μl , 42°C water bath for 50 minutes, 70°C water bath for 15 minutes to stop the reaction;
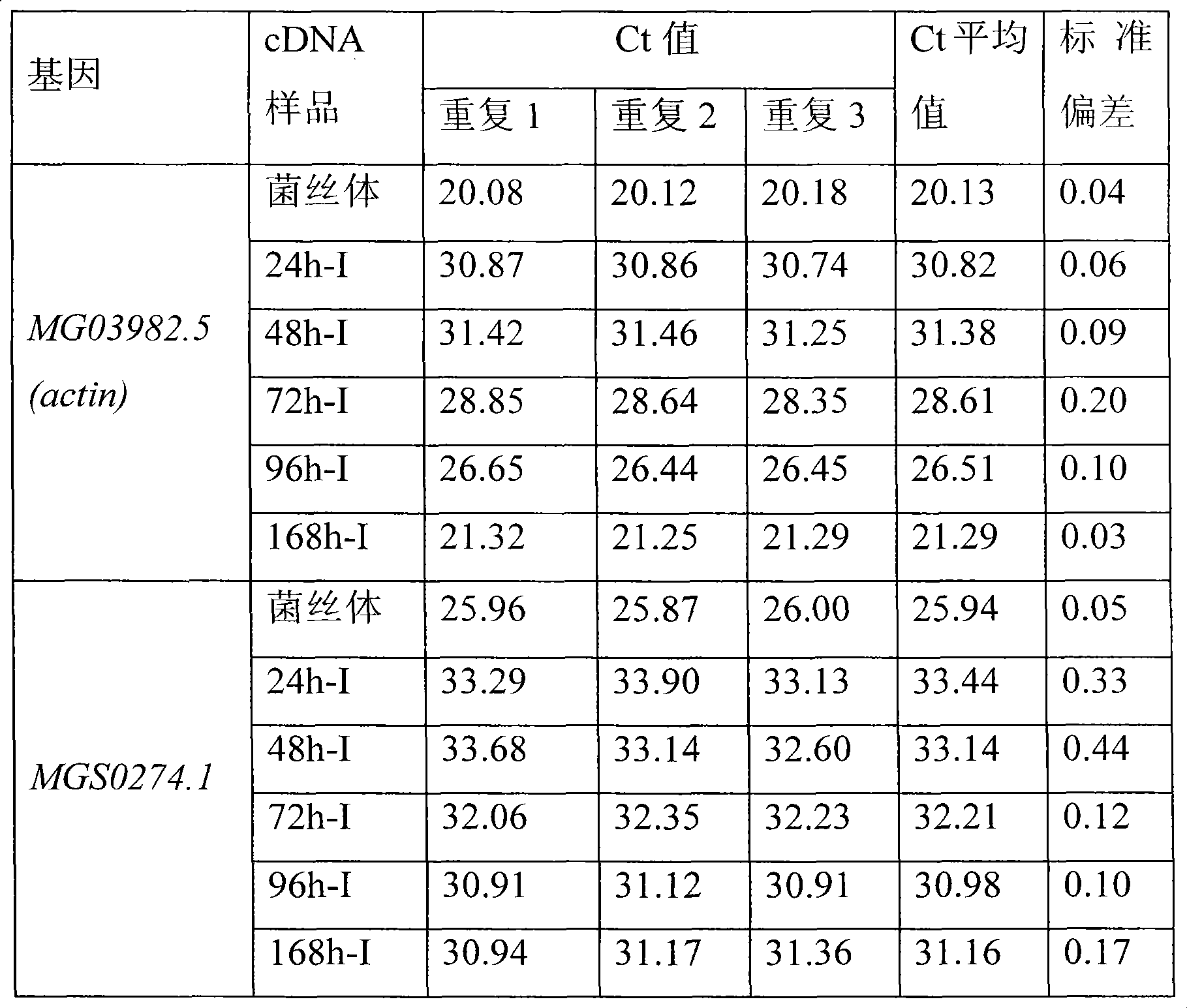
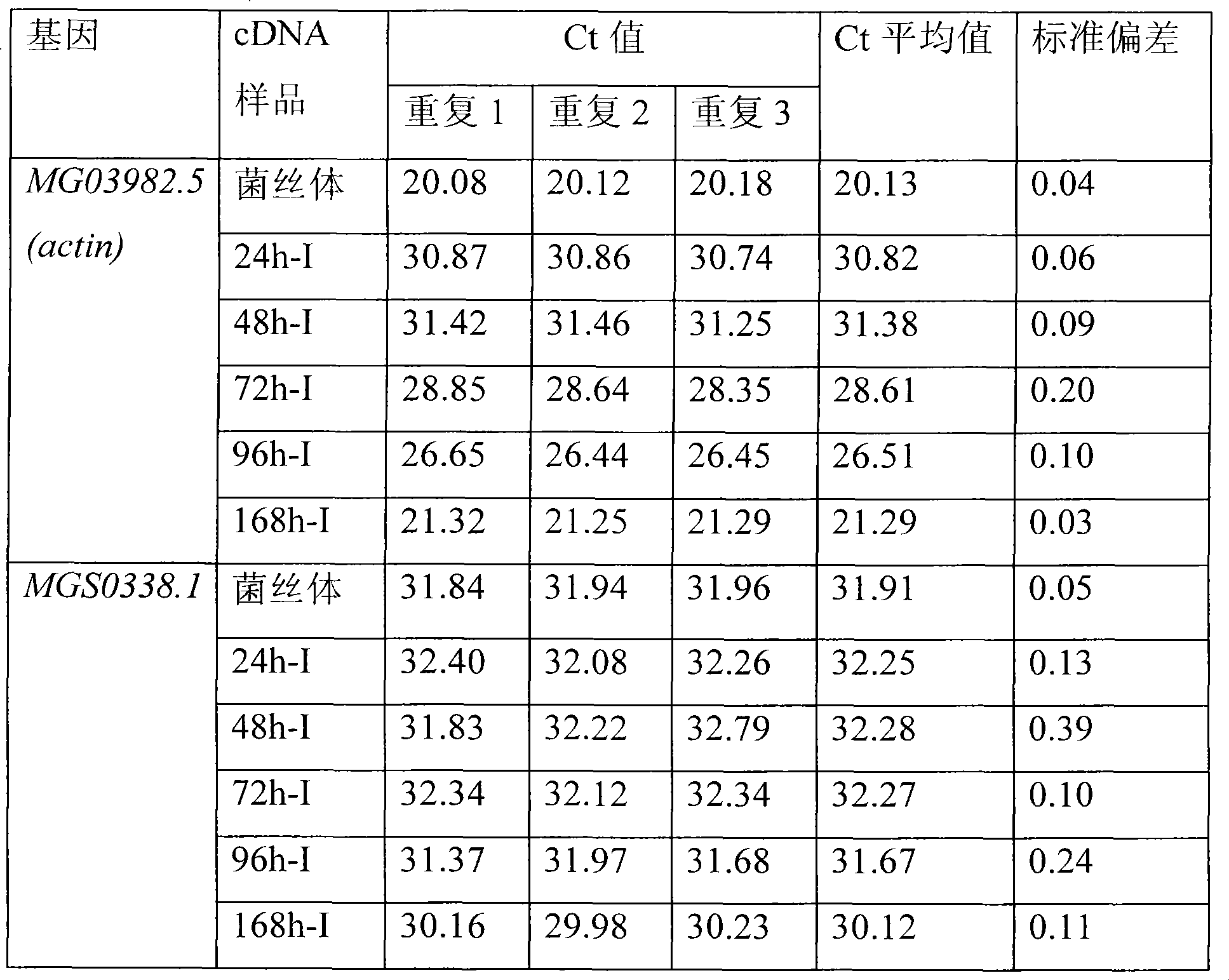
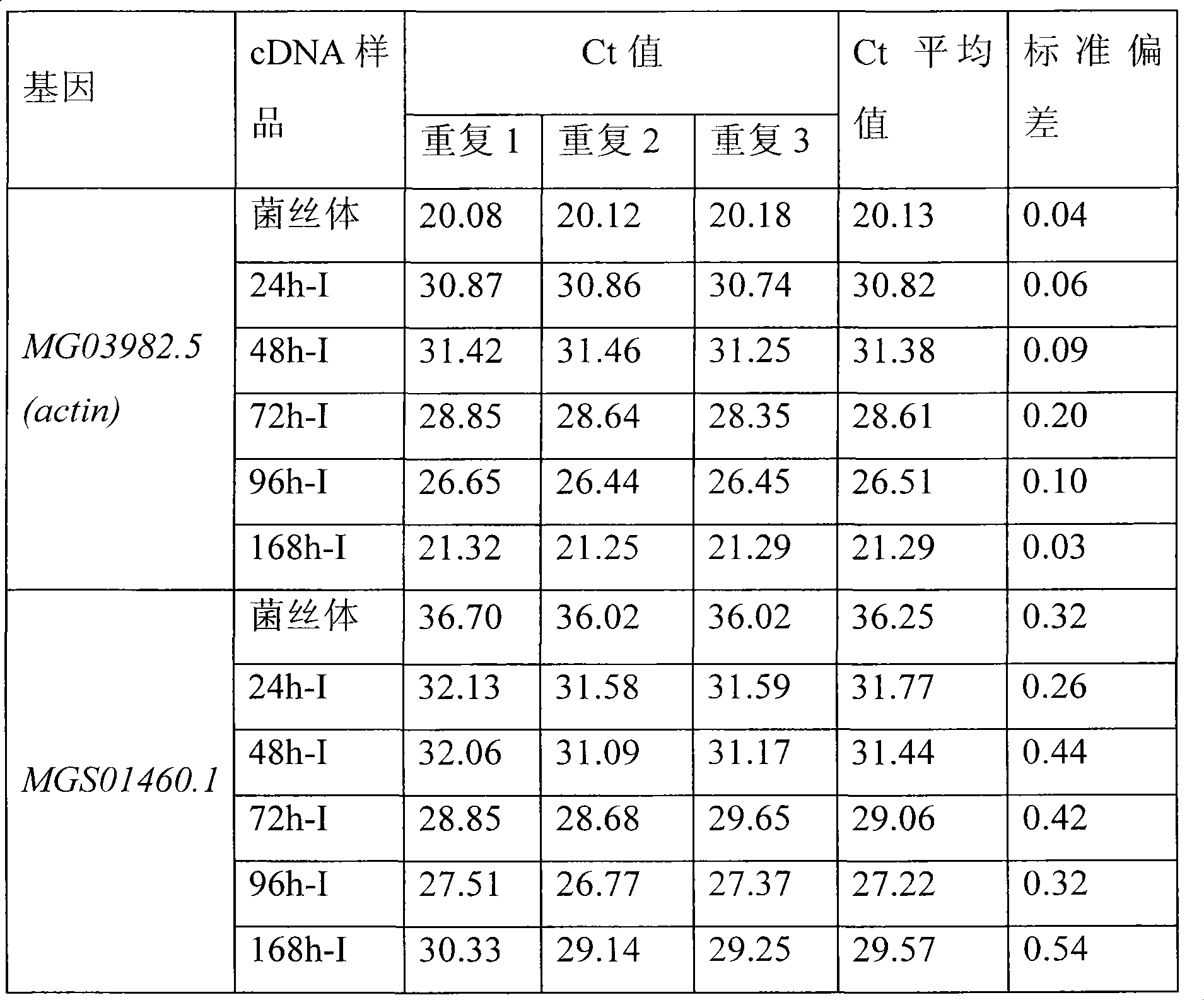
[0023] (4) Select the actin (actin) gene of Magnaporthe grisea MG03982.5 as an internal referen...
Embodiment 2
[0037] (1) culturing Magnaporthe grisea in liquid and extracting the total RNA of its mycelia, and reverse-transcribing the mRNA therein into cDNA;
[0038] (2) Inoculate the spores of Magnaporthe grisea on the leaves of rice susceptible variety Lijiang Xintuan Heigu, and extract the total RNA of the infected tissue at different time periods early after inoculation and reverse transcribe the mRNA therein into cDNA;
[0039](3) Synthesis of cDNA: Take 2.5 μg of each total RNA sample, add 0.5 μg / μl Oligo (dT) 2.5 μl, 10 mM dNTP 2.5 μl, RNase-free water to 30 μl, 65 ° C water bath for 5 minutes, and quickly place in Cool on ice, add 5×First-Strand Buffer 10μl, 0.1M DTT (dithiothreitol) 5μl, 40U / μl RNase inhibitor 2.5μl, 42℃ water bath for 2μmin, then add SuperScript II reverse transcriptase 2.5μl , 42°C water bath for 50 minutes, 70°C water bath for 15 minutes to stop the reaction;
[0040] (4) Select the actin gene of Magnaporthe grisea MG03982.5 as an internal reference gene, ...
Embodiment 3
[0055] (1) culturing Magnaporthe grisea in liquid and extracting the total RNA of its mycelia, and reverse-transcribing the mRNA therein into cDNA;
[0056] (2) Inoculate the spores of Magnaporthe grisea on the leaves of rice susceptible variety Lijiang Xintuan Heigu, and extract the total RNA of the infected tissue at different time periods early after inoculation and reverse transcribe the mRNA therein into cDNA;
[0057] (3) Synthesis of cDNA: take 2.5 μg of each total RNA sample, add 0.5 μg / μl Oligo (dT) 2.5 μl, 10 mM dNTP 2.5 μl, RNase-free water to 30 μl, 65 ° C water bath for 5 minutes, quickly placed in Cool on ice, add 5×First-Strand Buffer 10μl, 0.1M DTT (dithiothreitol) 5μl, 40U / μl RNase inhibitor 2.5μl, 42℃ water bath for 2μmin, then add SuperScript II reverse transcriptase 2.5μl , 42°C water bath for 50 minutes, 70°C water bath for 15 minutes to stop the reaction;
[0058] (4) Select the actin gene of Magnaporthe grisea MG03982.5 as an internal reference gene, an...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com