Obtaining method and use of novel oncolytic adenovirus construct with selective tumor blockage STAT3
A technology for constructing and recombining adenovirus, which is applied in the field of d5 E3 region 29477-297, can solve the problems of lack of tumor specificity, restricting tumors, weak efficiency of adenovirus in lysing tumor cells, etc., and achieves increased amplification, copying The effect of number increase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
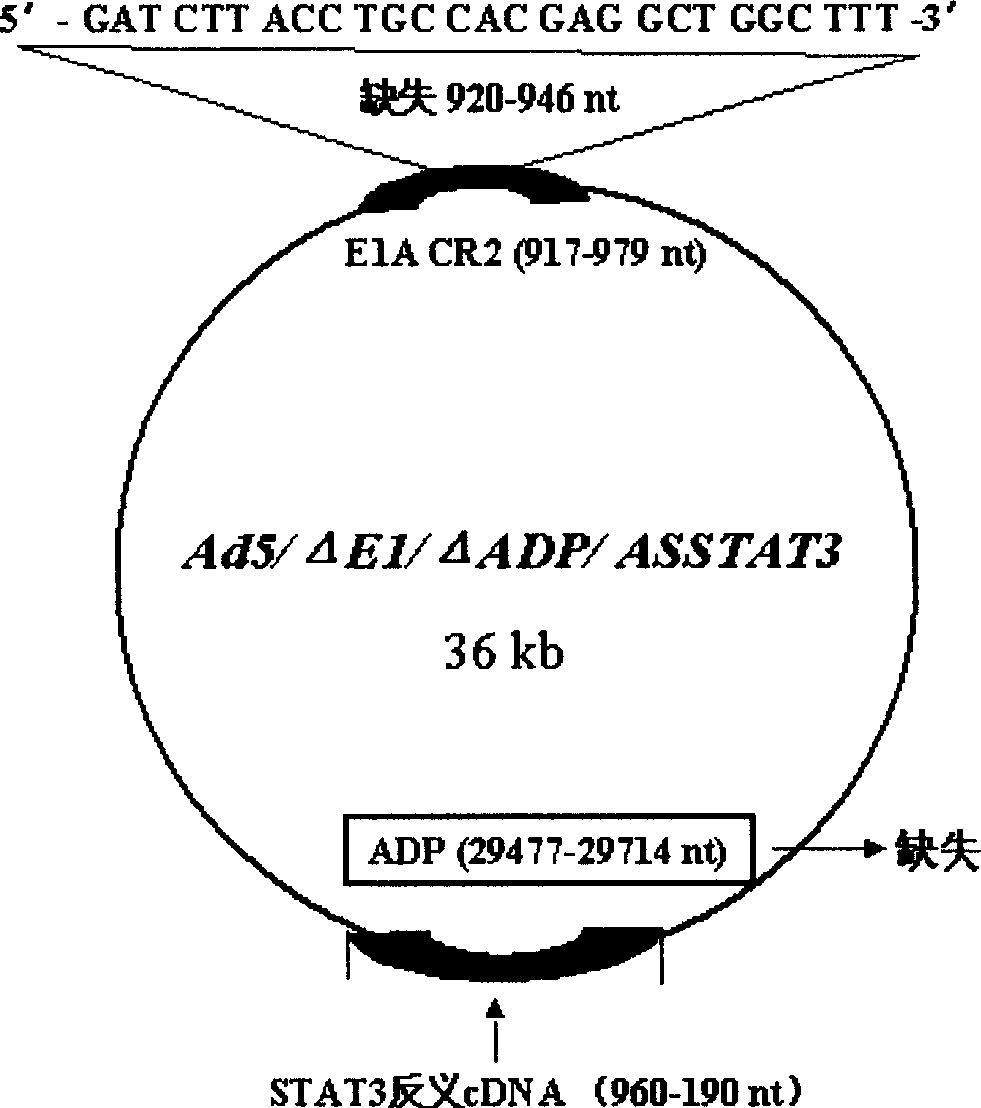
[0024] Example 1. Construction of pXC1 series mutants (Δ920-946pXC1)
[0025] pXC1 was purchased from Microbix Biosystem Inc. (Toronato, Ontario, Canada, catalog number: PD-01-03), and this plasmid contains the 21-5790nt sequence of human adenovirus type 5 (Ad5).
[0026] Deletion of 920-946nt by 3 times PCR method, acquisition of fragment 1: primer 1=5′-CG GGA TCC GGG CCCCCA TTT CC-3', equivalent to 9883-9902nt, the underlined part is the BamHI restriction site; primer 2 = 5'- GTC ACT GGG TGG ATC GAT CAC CTC CGG TAC-3', corresponding to 922-905nt, the underlined part is complementary to primer 3;
[0027] Use pXC1 as a template for PCR reaction, the total volume of the reaction system is 100 μl, including:
[0028] Contains MgCl 2 10 μl of 10x PCR buffer
[0029] 2mM dNTP 10μl
[0030] 10 μM Primer 1 1 μl
[0031] 10 μM Primer 2 1 μl
[0032] pXC1 10ng / μl 1μl
[0033] pfu high fidelity Taq enzyme 2.5u
[0034] Add water to 100μl;
[0035] The reaction condition...
example 2
[0041] Example 2. Construction of Δ920-946 Ad5 recombinant adenovirus
[0042] pBHGE3 was purchased from Microbix Biosystem Inc. (Toronato, Ontario, Canada, catalog number: PD-01-12), this plasmid contains the entire genome sequence except the ADd5 packaging signal (194-358nt).
[0043]When pBHGE3 is obtained from Microbix Biosystem Inc., the total amount is 10 μg. First, electroporate into competent bacteria, pick positive clones, and extract plasmids. The obtained plasmids are treated with CsCl 2 -EB purified by ultracentrifugation.
[0044] Homologous recombination method to obtain Δ920-946 Ad5 recombinant adenovirus construct, the method is as follows:
[0045] Plant 7.5×10 in a 15cm petri dish 5 293 cells, the culture medium is 10% FBS DMEM, by the next day, the cells should be 1-1.5×10 6 , about 70% of the cells are confluent; 3-4 hours before transfection, replace with fresh culture medium.
[0046] Prepare co-transfection DNA-calcium phosphate solution: 1600 μl st...
example 3
[0061] Example 3. Construction of subcloning vector pCDNA3.1-ΔADP of Ad5 E3 region
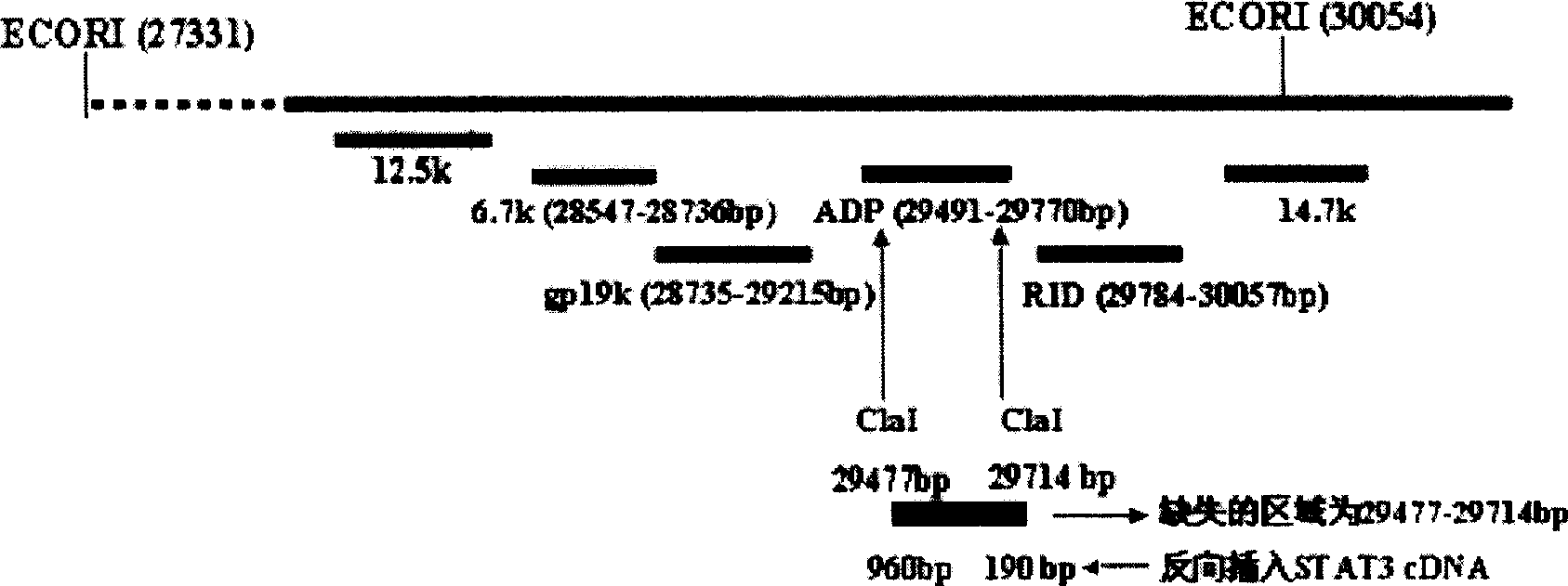
[0062] The Ad5 E3 region is called the Ad5 early region 3. Driven by the endogenous promoter, this region encodes 7 proteins. For the sequence, structure and function, see Figure 1: 12.5k, 27858-28179nt, the function is unknown; 6.7k, 27547-28736nt, together with the RID complex, inhibits the expression of TRAIL receptors 1 and 2 on the cell surface; gp19k, 28735-29215nt, binds to MHC class I antigens, inhibits its presentation to the cell surface, and escapes the clearance of CTL; ADP, 29419 -29770nt, lyses cells and releases virus; RIDα, 29784-30057nt, forms a complex with RIDβ, prevents the lysis of TNF, and clears FAS antigen; RIDβ, 30062-30458nt and 14.7k, 30453-30837, inhibits the lysis of TNF.
[0063] The purpose of this experiment is to delete the 29477-29714nt region of the E3 region and insert an exogenous therapeutic gene into the E3ADP region of the recombinant adenovirus.
[006...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com