Oligonucleotide and uses thereof
An oligonucleotide, mir-21 technology, applied in DNA/RNA fragmentation, microbial determination/test, recombinant DNA technology, etc., can solve the problem of no significant improvement, poor curative effect, and low survival rate of middle-advanced patients. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] The extraction of embodiment 1 total RNA and miRNA (mirVana TM microRNA extraction kit, Ambion company)
[0058] 1 Add 600μl Lysis / Binding Solution to lyse the tissue;
[0059] 2 Add microRNA Homogenate Additive at 1 / 10 the volume of the lysate, mix well, and place on ice for 10 minutes;
[0060] 3 Add the same volume of phenol:chloroform solution as the lysate (without adding microRNA Homogenate Additive), mix well, and centrifuge at the maximum speed (10,000×g) for 5 minutes at room temperature;
[0061] 4 Transfer the supernatant to a clean tube and note down the volume of the liquid;
[0062] 5 Add 1 / 3 volume of 100% alcohol to the supernatant and mix thoroughly;
[0063] 6 Add the lysate / alcohol mixture to the Filter Cartridge column, centrifuge at ~10,000×g for 15s, and collect the filtrate;
[0064] 7. The main thing retained on the Filter Cartridge column is macromolecular RNA, and the retained RNA is recovered according to the standard procedure of total R...
Embodiment 2
[0072] Preparation of embodiment 2cDNA (miRNA reverse transcription):
[0073] The miRNA extracted in Example 1 was reverse-transcribed to prepare a cDNA template (reverse transcription primer sequences are shown in Table 1).
[0074] A: Reverse transcription reaction system:
[0075] Reagent name (both buffer and enzyme are
promega company)
Dosage / tube
5×Reverse Transcription buffer
(buffer)
4ul
RT Primer Mix (1uM) (primer mixture)
1.25ul
dNTP (10mM) (four deoxyribonucleotides
Mixture)
0.75ul
RNA (template)
2ul
RTase (200U / ul) (reverse transcriptase)
0.5ul
DEPC H2O
to 20ul
[0076] B: Reverse transcription reaction conditions:
[0077] The reaction conditions are: 16°C for 30 minutes; 42°C for 30 minutes; 85°C for 10 minutes.
Embodiment 3
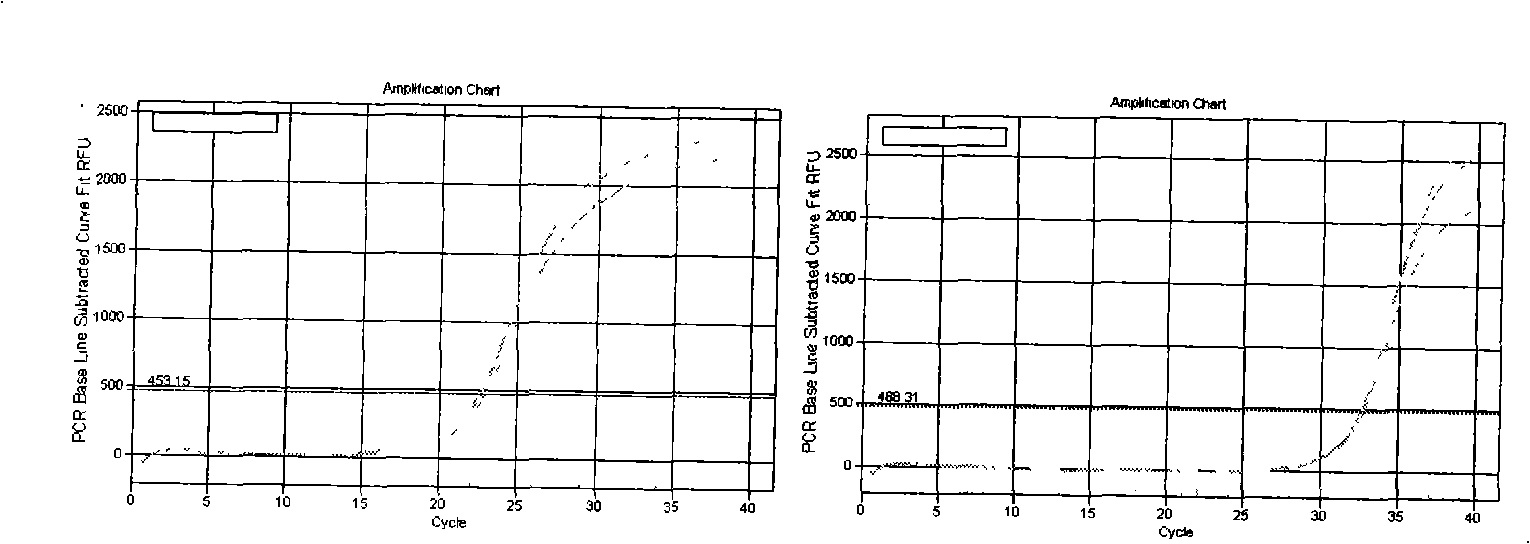
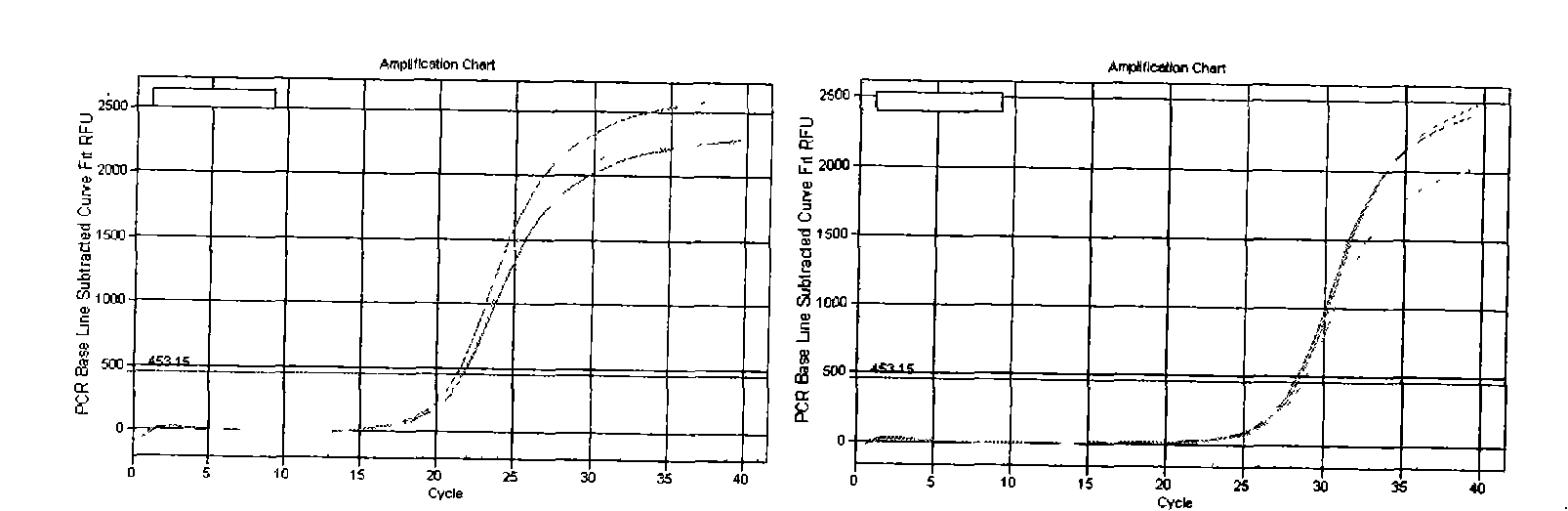
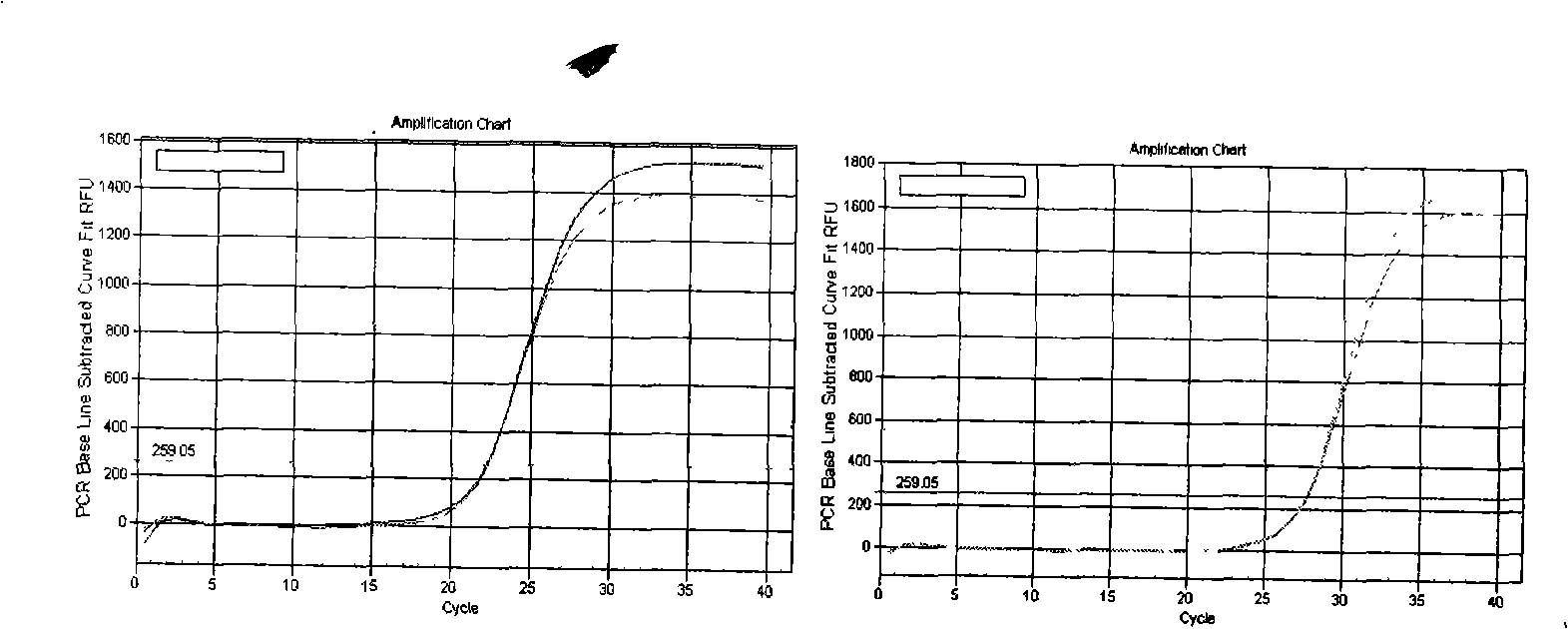
[0078] Embodiment 3 Dye method fluorescent quantitative PCR detection
[0079] A: Dilution of cDNA template:
[0080] The cDNA obtained after reverse transcription in Example 2 was diluted 3 times, for example: add 40 μl RNase / DNase free ddH2O to 20 μl system, and mix well.
[0081] B: Dye method fluorescent quantitative PCR system:
[0082] Reagent name
Dosage / tube
2×Real-time PCR Master Mix
10μl
PCR Forward Primer (10uM)
0.2μl
[0083] PCR Reverse Primer (10uM)
0.2μl
DNA / cDNA Template
2μl
Taq DNA polymerase (5U / μl)
0.2μl
dd H 2 o
To 20μl
[0084] Among them, the primers used by U6 are F Primer: ATTGGAACGATACAGAGAAGATT; R Primer: GGAACGCTTCACGAATTTG.
[0085] The primers used for miR-21 detection are:
[0086] First group:
[0087] RT Primers:
[0088] 5’-GTCGCAATCCATGAGAGATCCCTACCGACATGGATTGCGACTCAACA
[0089] Forward Primer: 5'-GCGGTAGCTTTATCAGACTGATGT-3'
[0090] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com