Method for preparing nucleotide by reaction separation coupling technology
A reaction separation coupling and nucleotide technology, applied in the field of biocatalysis and bioseparation, can solve the problems of yield reduction, achieve the effects of reducing energy consumption, eliminating product inhibition, improving enzyme utilization and reaction yield
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
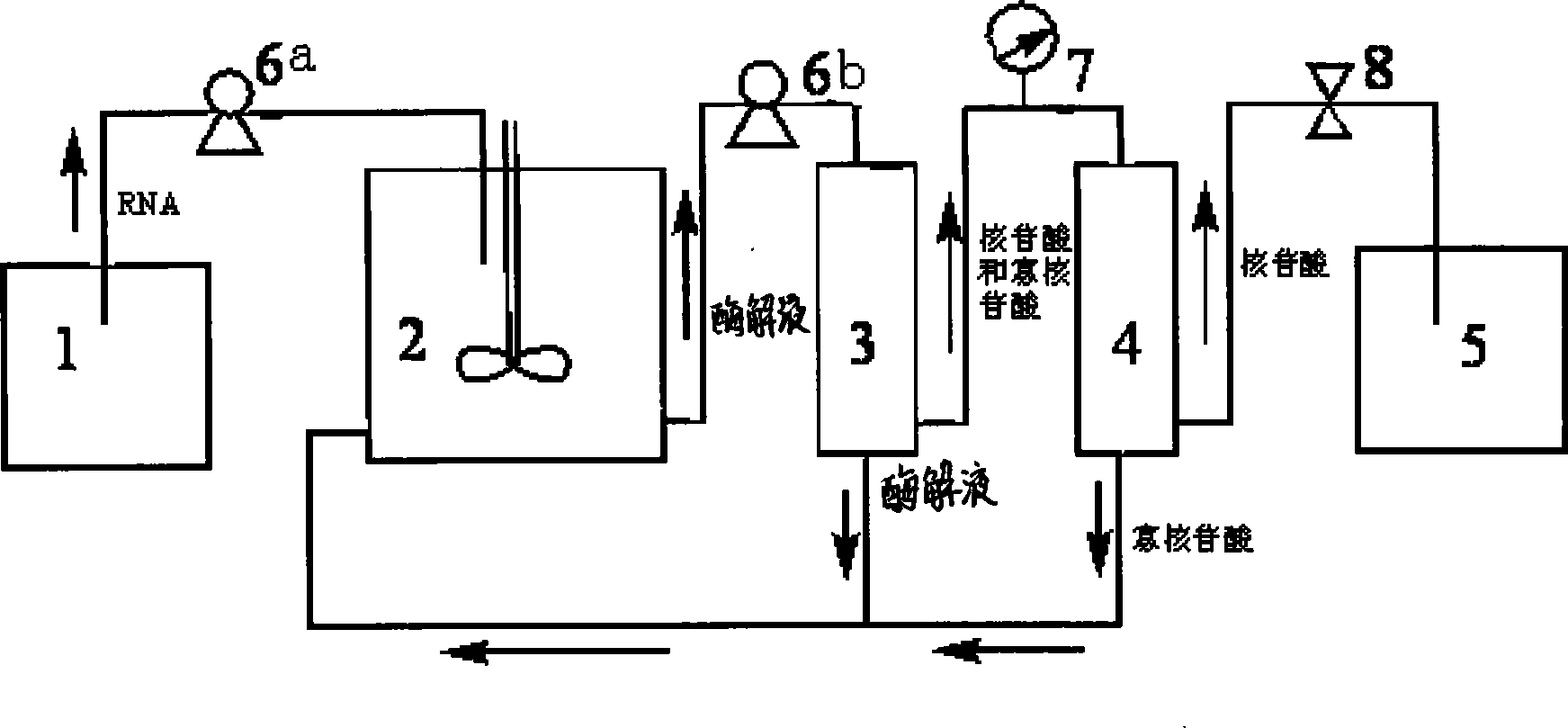
[0026] Put the nuclease P1 with a concentration of 3000μ / mL into a 5L enzymatic hydrolysis reactor, and use a peristaltic pump to continuously pump the RNA solution with a concentration of 20g / L and pH5.0 to carry out the enzymatic hydrolysis reaction, and control the enzymatic hydrolysis temperature to 70°C , the RNA solution was pumped in at a flow rate of 20 mL / min. After 1 hour of reaction, start to pump the reaction liquid into the polysulfone hollow fiber ultrafiltration membrane for ultrafiltration. 2 . The ultrafiltrate flows into the polysulfone hollow fiber nanofiltration membrane for nanofiltration separation. The molecular weight cut-off of the nanofiltration membrane is 1000 Daltons, and the filtrate is nucleotides, which flow into the storage tank for collection. The ultrafiltration and nanofiltration residues flow back into the reactor to continue enzymatic hydrolysis. The total reaction time is 4 hours, the final RNA hydrolysis rate reaches 96%, and the nucle...
Embodiment 2
[0028] Put nuclease P1 with a concentration of 3000μ / mL into a 50L reactor, and use a peristaltic pump to continuously pump an RNA solution with a concentration of 50g / L and pH 6.0 to carry out the enzymatic hydrolysis reaction. The solution was pumped in at a flow rate of 50 mL / min. After one hour of reaction, start to pump the reaction liquid into the polysulfone hollow fiber ultrafiltration membrane for ultrafiltration. The molecular weight cut-off of the ultrafiltration membrane is 7000 Daltons, the flow rate is 70mL / min, and the pressure does not exceed 0.5Kg / cm 2 . The ultrafiltrate then flows into the polysulfone hollow fiber nanofiltration membrane for nanofiltration separation. The molecular weight cut-off of the nanofiltration membrane is 300 Daltons, and the filtrate is nucleotides, which flow into the storage tank for collection. The ultrafiltration and nanofiltration residues flow back into the reactor to continue enzymatic hydrolysis. The total reaction time is...
Embodiment 3
[0030] Put nuclease P1 with a concentration of 3500μ / mL into a 100L reactor, and use a peristaltic pump to continuously pump an RNA solution with a concentration of 100g / L and a pH of 7.0 for enzymolysis reaction. The enzymolysis temperature is controlled at 75°C. The solution was pumped in at a flow rate of 70mL / min. After one hour of reaction, pump the reaction liquid into the polysulfone hollow fiber ultrafiltration membrane for ultrafiltration. The molecular weight cut-off of the ultrafiltration membrane is 7000 Daltons, the flow rate is 100mL / min, and the pressure does not exceed 0.5Kg / cm 2 . The ultrafiltrate flows into the polysulfone hollow fiber nanofiltration membrane for nanofiltration separation. The molecular weight cut-off of the nanofiltration membrane is 400 Daltons, and the filtrate is nucleotides, which flow into the storage tank for collection. The ultrafiltration and nanofiltration residues flow back into the reactor to continue enzymatic hydrolysis. The ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com