Method for detecting the polymorphism of ADH2 genes
A gene polymorphism and gene technology, applied in the field of ADH2 gene polymorphism detection, can solve the problems of increased dosage, high price, instability, etc., and achieve the effect of easy acquisition, smooth amplification, and clear bands
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] 1. Sequence search and polymorphic site determination
[0032] According to NCBI's GENE and SNP databases, the full sequence and polymorphic site information of the ADH2 gene were obtained, and the polymorphic sites to be studied were determined. The full sequence of the ADH2 gene and the information on the ADH2 polymorphic site are obtained from the following webpage:
[0033] http: / / www.ncbi.nlm.nih.gov / entrez / viewer.fcgi? db=nuccore&id=89161207 ,
[0034] http: / / www.ncbi.nlm.nih.gov / SNP / snpref.cgi? rs=1229984 ,
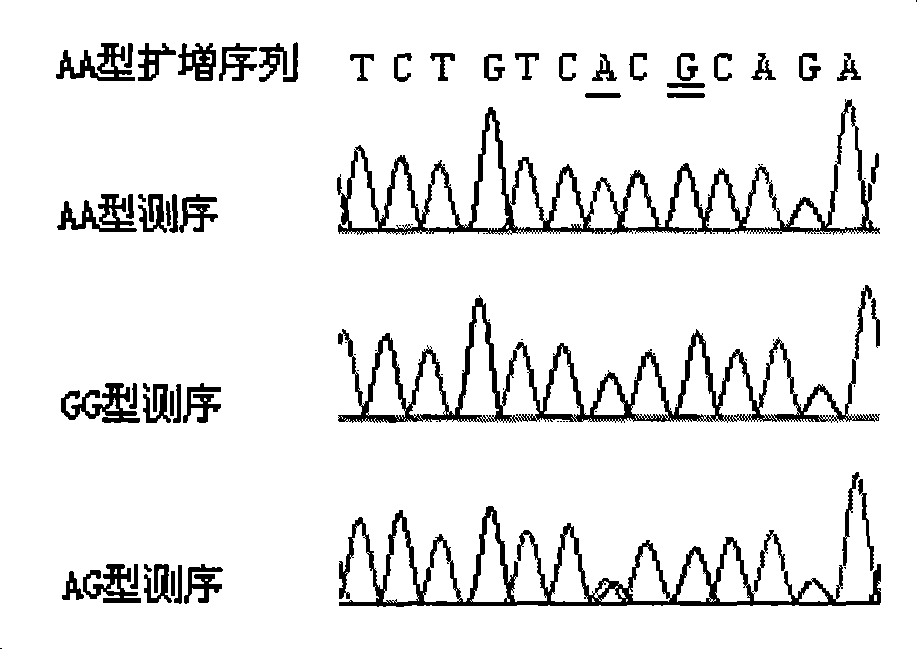
[0035] According to the information on the above web page and relevant documents, the present invention determines that the ADH2 polymorphic site is A / G polymorphic (rs1229984), located at chromosome 4, exon 3, genome position 3240.
[0036] 2. Sequence analysis and primer design
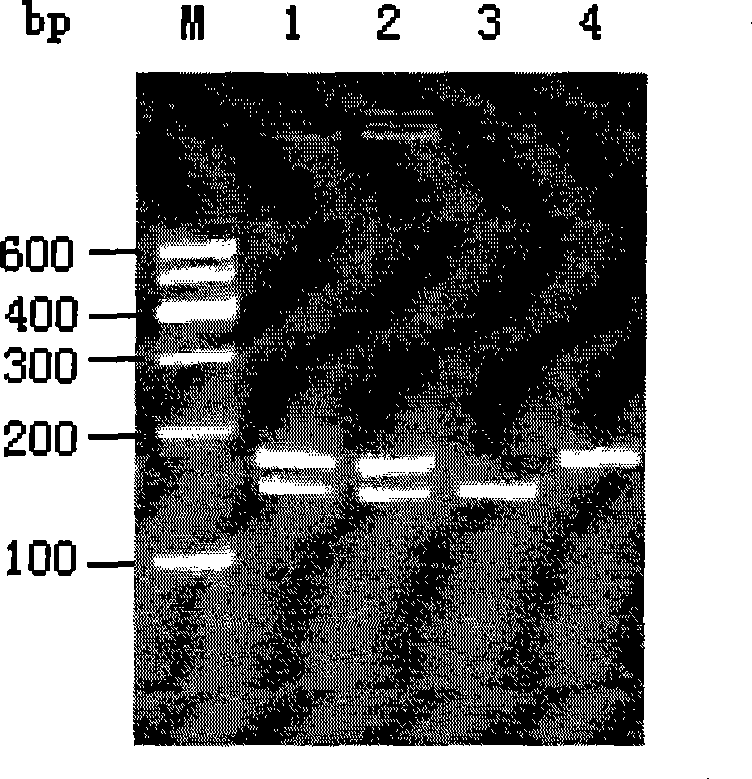
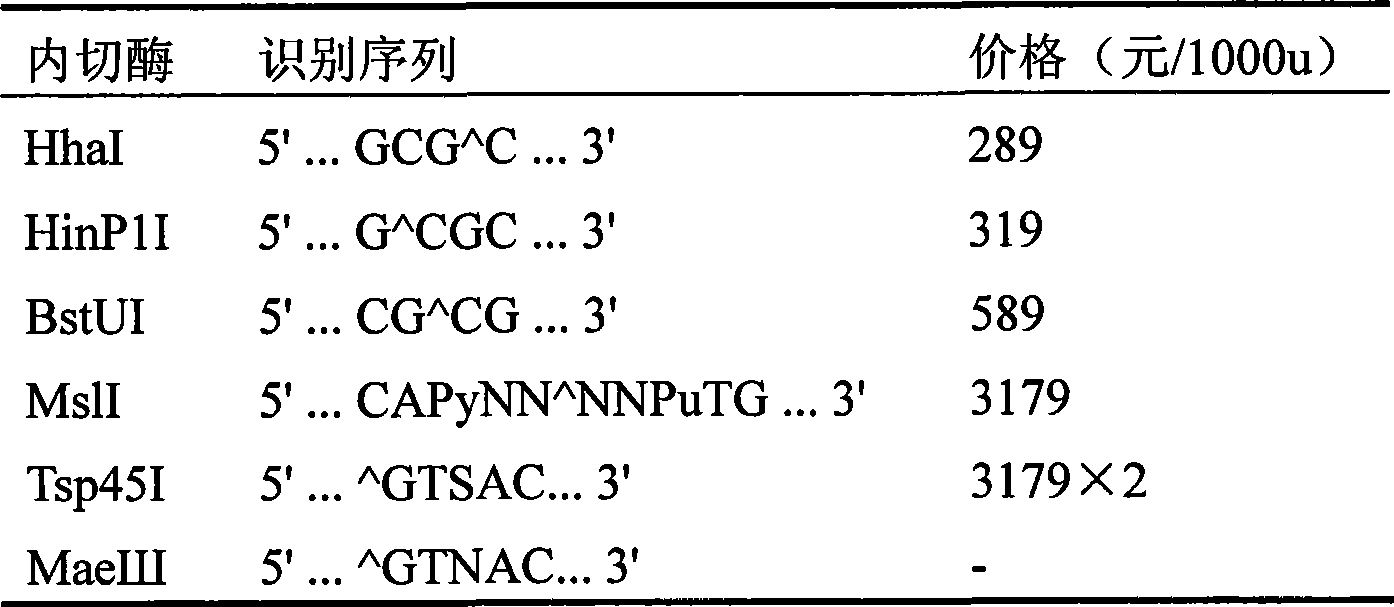
[0037] According to sequence analysis, the A / G polymorphism can be recognized by three endonucleases (and its isozymes) of Tsp45I, Ms1I and MaeIII, that is, the A allele ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com