Kit for quickly extracting plant genome and applications thereof
A plant genome and kit technology, applied in the field of molecular biology, can solve the problems of demanding samples, and achieve the effect of simple method, low reagent cost and good safety
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
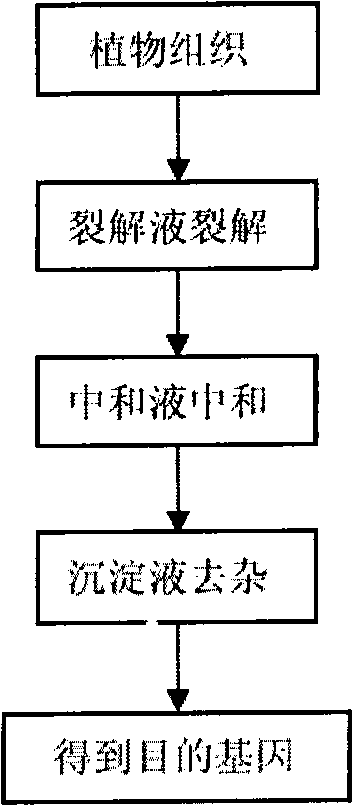
[0054] Embodiment 1 uses the method of the present invention to extract various plant genomes
[0055] As a rapid preparation method of plant genome for molecular detection, we hope that this method has wide applicability and can be widely used in the molecular detection of genetically modified products in the food field. Edible tissues or organs in plants can be roughly divided into the following categories: roots, stems, leaves, flowers, fruits, and seeds. In order to test the universality of the method, we must not only cover edible tissues of plants when selecting samples Partly, try to select plants of different families and genera as research objects. Therefore, when selecting samples, this example selects (1) cereals: corn, soybeans, peanuts; (2) stems; Chrysanthemum chrysanthemum, celery; (3) Leaves: rapeseed; (4) roots: carrots; (5) flowers: broccoli; (6) fruits: tomatoes.
[0056] The specific operation is as follows:
[0057] a. Cereals: BT176 transgenic corn and ...
Embodiment 2
[0080] Example 2 Using the method of the present invention to amplify target genes of different fragment sizes
[0081] The plant genome obtained by the method of the present invention presents a diffuse state due to degradation, and the genome electrophoresis bands mainly focus on 1000bp to 200bp, while the genes amplified in the above examples are all about 200bp plant internal standard genes, so there is better amplification. increase the effect. In order to compare the effect of this method on the amplification of genes with different fragment sizes, in this example, three genes in BT176 corn were selected as the amplification objects. The specific operations are as follows:
[0082] The BT176 transgenic corn was crushed with a grinder and passed through a 60-mesh sieve.
[0083] According to the method of the present invention of embodiment 1 and CTAB method (CTAB method is referring to: Xu Fang, Li Xin, Luo Xin, Liu Zhiguo. Qualitative PCR detection [J] of genetically mod...
Embodiment 3
[0088] The detection result of the sensitivity of embodiment 3 the present invention
[0089] After mixing BT176 transgenic corn flour and soybean flour at a mass ratio of 50%, 10%, 1%, and 0.1%, the genome was extracted according to the method of the present invention in Example 1, and the extracted DNA was used as a template for PCR electrophoresis detection. The maize internal standard gene IVR226 was selected as the primer for PCR detection, and the target fragment amplified by the reaction was about 226bp: the base sequence of the primer IVR226 was:
[0090] F5′-CCG CTG TAT CAC AAG GGC TGG TCA C-3′
[0091] R5′-GGA GCC CGT GTA GAG CAT GAC GAT C-3′
[0092] The PCR reaction system, 10×PCR buffer, PCR reaction conditions and detection conditions are all consistent with Example 1.
[0093] The results show that: when the content of corn flour is less than or equal to 1%, the target gene bands that can be distinguished by naked eyes can be obtained after PCR amplification, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com