Proteinaceous pharmaceuticals and uses thereof
A protein and cysteine technology, which is applied to peptide/protein components, pharmaceutical formulations, medical preparations containing active ingredients, etc., can solve the problems of no definite folding, no single stable structure, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0912] Example 1: Randomization of CDP 6_6_12_3_2
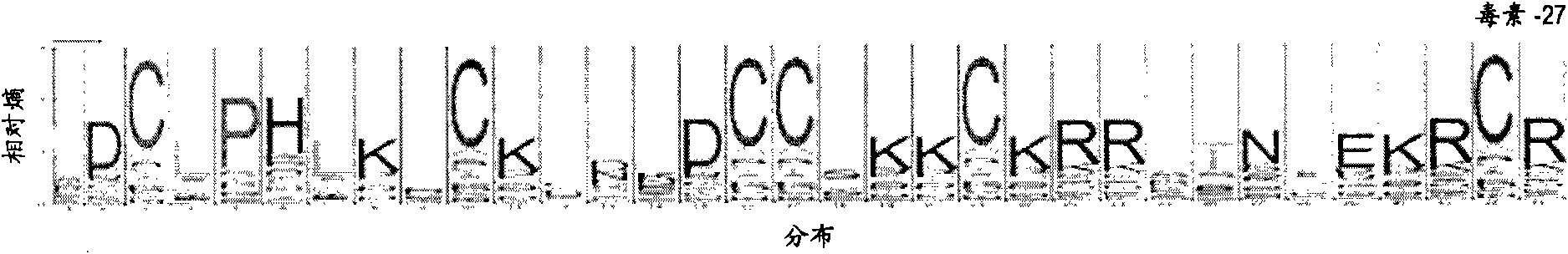
[0913] The following examples describe the design of CDP 6_6_12_3_2 based libraries. Partial sequences matching CDP 6_6_12_3_2 were searched in the TrEMBL protein sequence database. A total of 71 sequences matched this CDP. Calculate the amino acid abundance for each site as shown in Table 5. For each non-cysteine site, we selected a randomization scheme based on the following criteria: a) avoiding the introduction of stop codons, b) avoiding the introduction of additional cysteine residues, c) allowing for >3% specific position where a large number of amino acids were observed, d) minimized the incorporation of amino acids not observed in any of the 71 native sequences matching this CDP.
[0914]
[0915]
Embodiment 2
[0916] Example 2: Protein expression and folding in E. coli
[0917] The oligonucleotides were cloned into expression plasmid vectors driving protein expression in E. coli cytoplasm. A preferred promoter is T7 (Novagen pET vector series; Kan marker) in E. coli strain BL21 DE3. A preferred method of inserting these oligos is the modified Kunkel method (Scholle, D., Kehoe, JW and Kay, B.K. (2005) Efficient construction of a large collection of phage-displayed combinatorial peptide libary. Comb. Chem. & HTP Screening8: 545-551). A different approach is to perform 2-oligo PCR on (full or partial) vectors, followed by digestion of unique restriction sites in the ends of the oligo-derived fragments, followed by ligation of matching non-palindromic overhangs (effective fragment inner join). The third method is to assemble the insert from 2 or 4 oligos by overlapping PCR, digest restriction sites at the end of the assembled insert, and then ligate it into the digested vector. Tran...
Embodiment 3
[0919] Embodiment 3: Design steps of antifreeze protein
[0920] Purpose: Design libraries for antifreeze repeat proteins
[0921] Strategy: The starting sequence for library design was derived from antifreeze protein from Tenebrio molitor (Genbank accession number AF160494). This protein is known to express well in E. coli. Both crystal and NMR structures can be obtained. The protein is built from repeating units that form cylinders. The core of the structure lacks hydrophobic amino acids, but each repeat unit contains a disulfide bond and an invariant serine and alanine residue. The first two figures form a capping motif with three disulfide bonds. This capping motif is speculated to form a folded core. Thus, the first two repeat units generally remain unchanged during in vitro evolution. see Figure 127 .
[0922] Structural features of antifreeze proteins were analyzed in order to select exchange points and to discover glutamine residue sites for Scholle mutagenesi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| purity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com