Method for producing l-amino acid using bacteria belonging to the genus escherichia
A technology of amino acids and bacteria, applied in the biological field, can solve problems such as unclear functions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0104] Example 1: Cloning of b2682, b2683, b1242 and b3434 genes into pΔlacZ
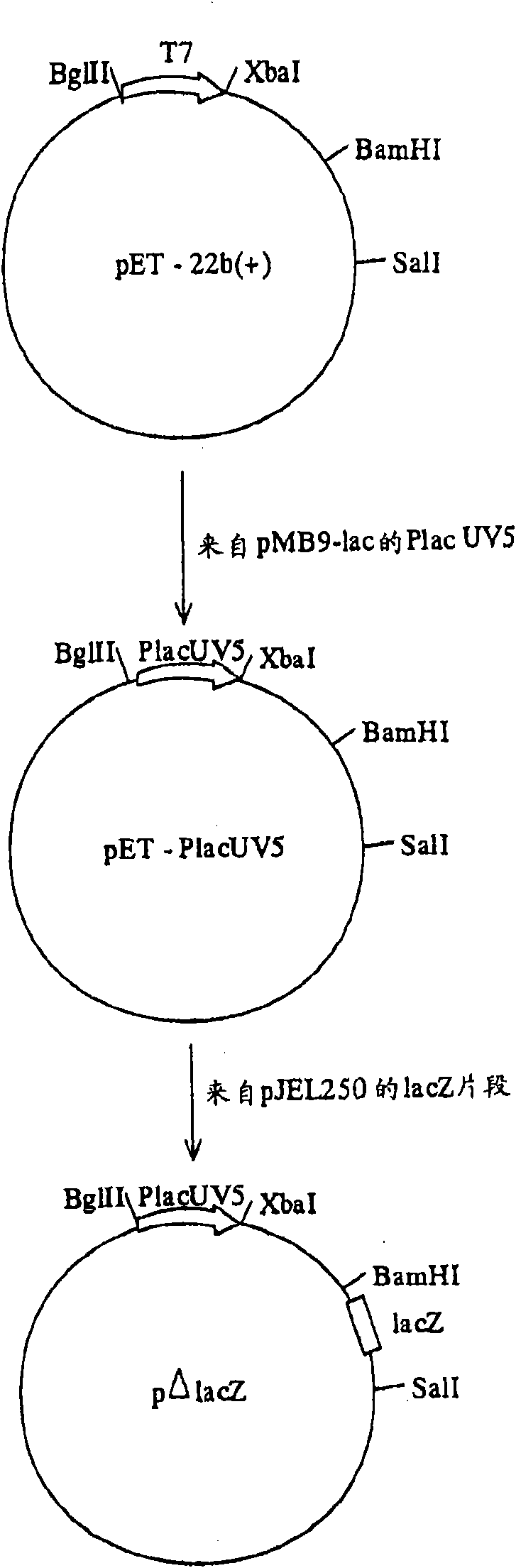
[0105] For cloning the b2682, b2683, b1242 and b3434 genes the pΔlacZ plasmid was used. Plasmid p[Delta]lacZ was derived from plasmid pET-22b(+) (Novagen, Madison. WI, USA). Treat the pET-22b (+) plasmid with enzymes BglII and Xba I, and treat it with the same endonuclease as the plasmid pMB9-lac (Fuller F., Gene, 19, 43-54, 1982) with P lac Fragment ligation of PCR products of the UV5 promoter. For amplified P lac The UV5 promoter fragment can use the PCR primers described in SEQ ID NO:7 and SEQ ID NO:8. The final plasmid added a partial structure of the lacZ gene (237 bp, no promoter) by cloning the Sal I-BamH I fragment of plasmid pJEL250 (Dymakova E. et al., Gene, 19, 43-54, 1982). The map of the obtained plasmid pΔlacZ is attached figure 1 Indicated.
[0106] The starting material for cloning the deduced reading frames (b2682 and b2683 genes) of Escherichia coli b2682 and b2683 was a PCR...
Embodiment 2
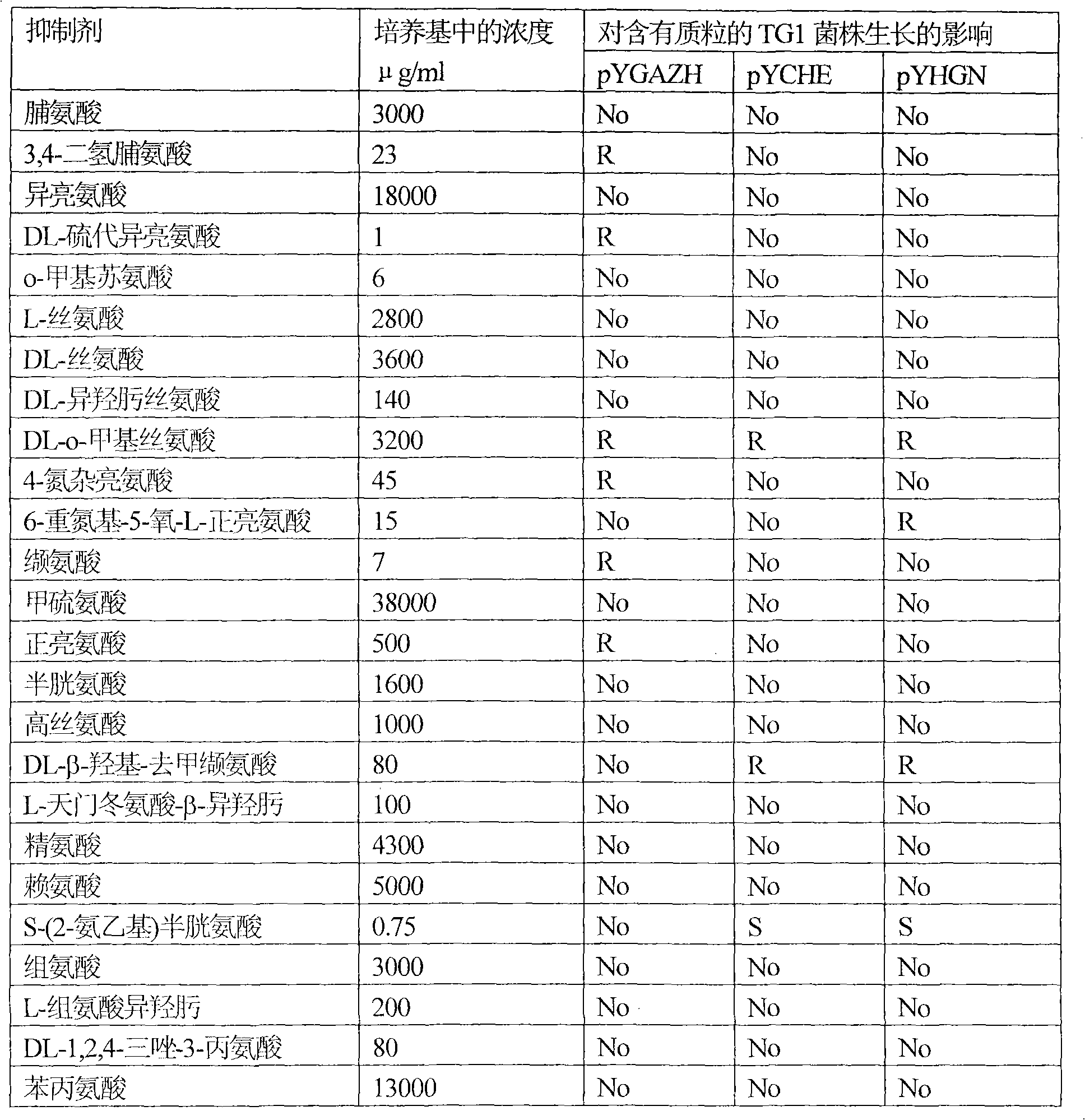
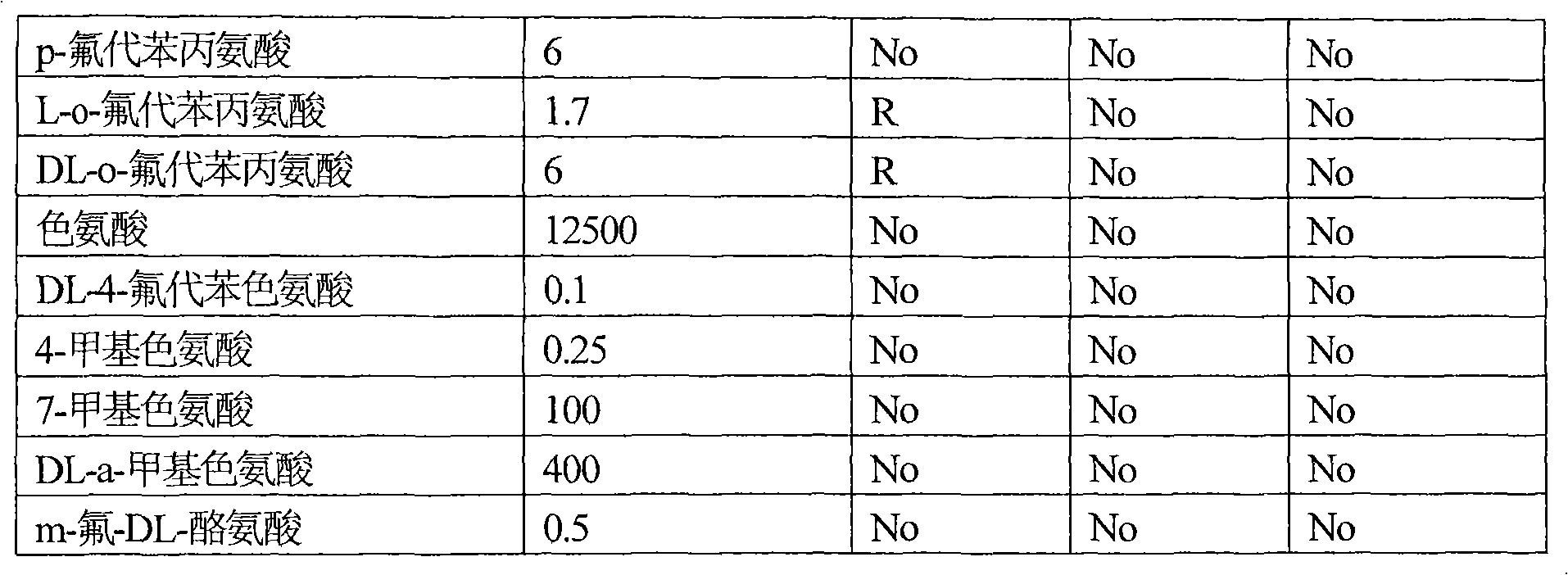
[0109] Example 2: Effects of the amplified b2862 and b2683 genes on the resistance of Escherichia coli strain TG1 to amino acids and their analogs.
[0110] Escherichia coli strains TG1(pYGAZH), TG1(pYCHE), TG1(pYHGN) and the TG1 strain containing the plasmid without insert (control strain) were cultured overnight on LB medium supplemented with 100 μg / ml ampicillin. Overnight cultures of all strains were diluted 25-fold with fresh LB medium supplemented with 100 μg / ml ampicillin and IPTG 0.5 mM, and incubated aerobically at 37° C. for 2 hours. Cultures in the logarithmic growth phase were diluted with 0.9% NaCl solution to inoculate approximately 1000 cells on Adams solid medium plates supplemented with 100 μg / ml ampicillin and IPTG 0.5 mM and amino acids or their analogs. After 2-4 days of incubation at 37°C, differences in the number and size of colonies formed between the TG1 strain harboring the hybrid plasmid and the control TG1 strain were recorded. The experimental res...
Embodiment 3
[0117] Embodiment 3: produce threonine with the bacterial strain that contains plasmid pYGAZH
[0118] The threonine-producing strain VL2054 was obtained by adding lac The plasmid pYGAZH of the b2682 and b2683 genes under the control of the UV5 promoter was transferred. The obtained strain was named VL2054(pYGAZH). Strain VL2054 is a derivative of strain VKPM B-3996, on the chromosome:
[0119] a) The target threonine operon is located at P R under the control of the promoter.
[0120] b) wild-type rhtA gene
[0121] c) Chromosomally silenced gene encoding transhydrogenase (tdh gene) and Tn5 (tdh::Tn5, Kan S ) on the silenced gene encoding kanamycin resistance (kan)
[0122] d) mutant ilvA 442
[0123] Bacterial strain VL2054 has been deposited (Russia 113545, Moscow, 1 Dorozhny proezd, 1) at Russian National Collection of Industrial Microorganisms (VKPM) on January 30, 2001, and the preservation number is VKPM B-8067, and was obtained in 2002 according to the Buda...
PUM
| Property | Measurement | Unit |
|---|---|---|
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com