Method for screening atherosclerosis related miRNA
A technology for atherosclerosis and screening methods, applied in the direction of biochemical equipment and methods, microbial measurement/testing, etc., to achieve the effect of perfecting the formation mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0018] Example 1 Isolation of Human Peripheral Blood Mononuclear Cells and Their Phenotypic Characterization After Different Times of Ox-LDL Inflammatory Stimulation
[0019] Heparin anticoagulated venous blood was taken from healthy adult volunteers, mononuclear cells were separated by density gradient centrifugation and adherence method, and monocytes were incubated together with ox-LDL (30mg / L) for 6 to 12 hours.
Embodiment 2
[0020] Example 2 Divided into monocyte group and ox-LDL inflammatory stimulation group at different time (6 hours, 12 hours), and Microarray analysis technology was used to detect the expression level of miRNAs in each group
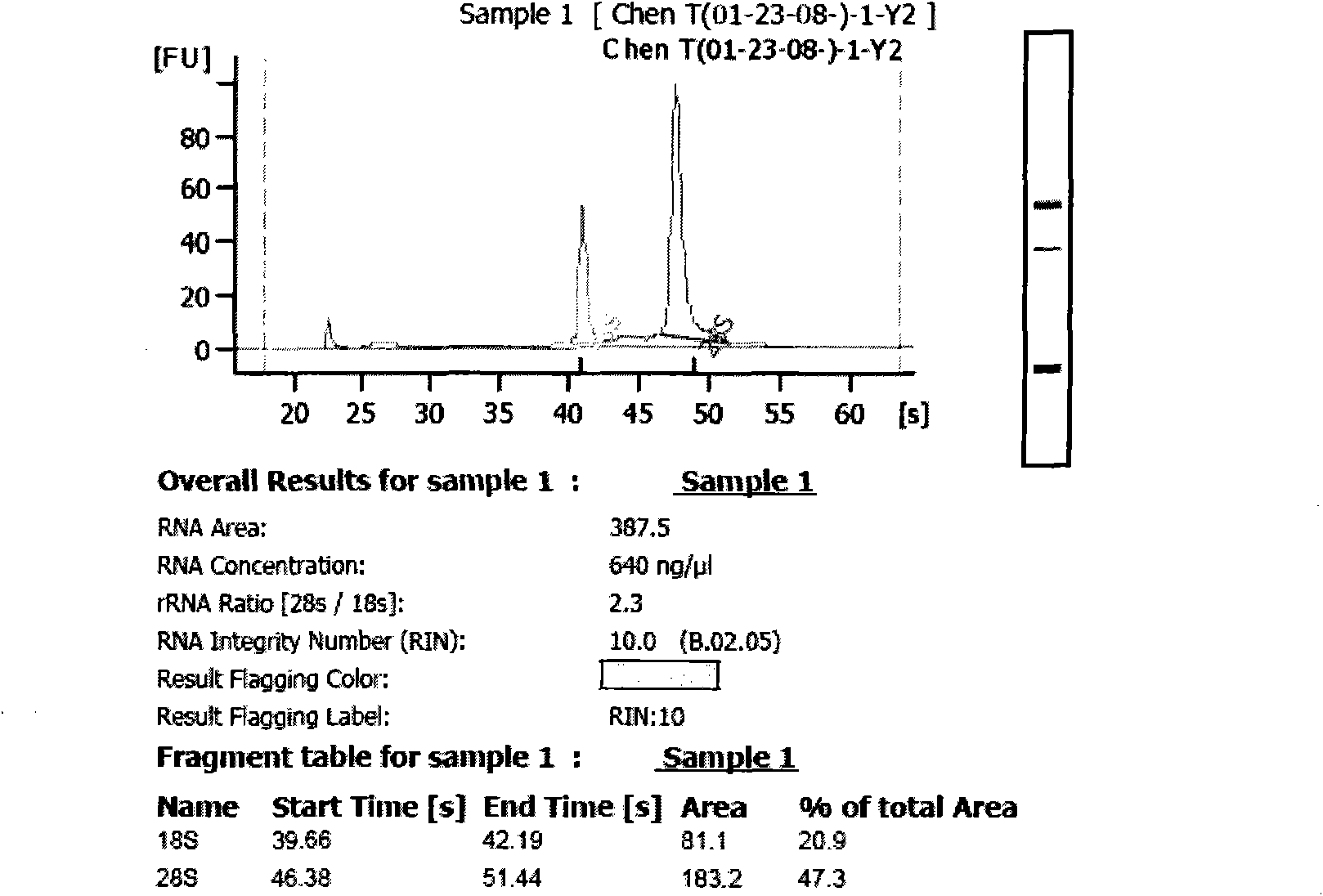
[0021] The Qiagen kit extracted more than 5 μg of total RNA samples, and separated them through YM-100 (Millipore) microcentrifuge filter columns to obtain small RNAs with fragments less than 300 nt. please see figure 1 As shown, the obvious 5S, 18S, and 28S electrophoresis bands can be seen from the figure, which proves that the small RNA is not lost.
[0022] Poly(A) polymerase adds a poly(A) tail to the 3′ end of the isolated small RNA, and then an oligonucleotide tag is ligated to the poly(A) tail for subsequent fluorescent labeling ( Cy3 and Cy5).
[0023] The labeled miRNAs were hybridized with the miRNAs chip: the hybridization reaction was performed overnight on the μParaflo microfluidic chip (Actic Technologies) by a microcirculation pump ...
Embodiment 3
[0024] Example 3 Detection confirms differential expression of miRNAs and predicts possible target mRNAs of corresponding miRNAs
[0025] The 5 miRNAs with the most significant difference between 6 and 12 hours of ox-LDL inflammatory stimulation screened by the chip were verified by fluorescent quantitative PCR on the 7000 reaction instrument with tagman probe (Applied Biosystems, Foster City, CA), and first used MuLV ( Multiscribe) reverse transcriptase and specific primers for reverse transcription, prepare a 15-μL reaction system according to the instructions, set cycle parameters at 16°C for 30 minutes, 42°C for 30 minutes, 85°C for 5 minutes, and stop temperature at 4°C. Then use RT products, Taqman 2X General PCR Master Mix and 5X MicroRNA Assay Mix containing primers and specific probes to prepare a 20-μL reaction system for amplification, set cycle parameters at 95°C for 10 minutes, 95°C for 15 seconds for denaturation and annealing amplification at 60°C for 60 seco...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com