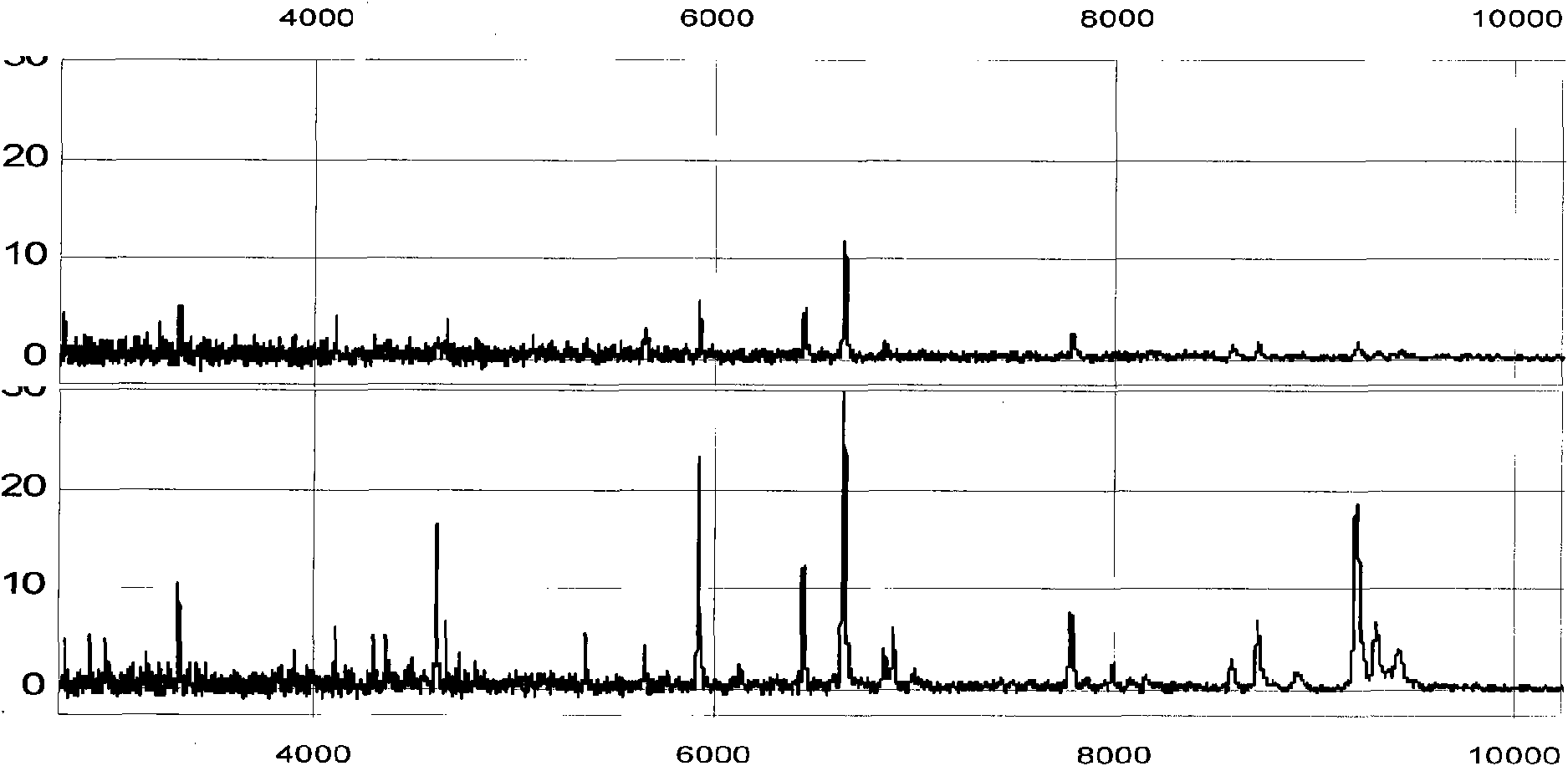
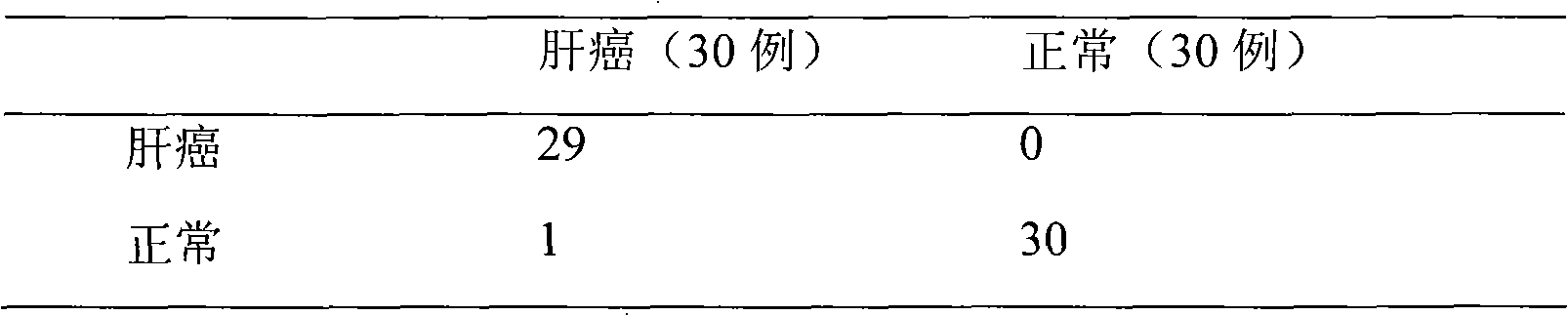
Method for analyzing proteome in biological sample
A biological sample and analysis method technology, applied in the preparation of test samples, analytical materials, biological testing, etc., can solve the problems of limited resolution, difficult to distinguish, limited gel capacity and sensitivity, etc., to improve the detection rate , early diagnosis of cancer, the effect of increasing the difference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Diagnosis of liver cancer
[0041] 1. Establish a standard fingerprint library of normal serum, the method is as follows:
[0042] 1,000 healthy volunteers with three years of continuous physical examination, half male and half male, aged 20 to 50, no family history, at least two generations of cancer-free patients, a few drops of blood were drawn, and the serum was separated. Add 1 μL of serum to 2 times the volume of U9 buffer to dilute (containing 9mol / L urea, 2% CHAPS, 50mmol / L Tris hydrochloric acid, 1% DTT, pH9.0), shake in ice bath for 30min, and make the proteome interact with urea. Then, 3 μL of 40 mM sodium acetate binding reagent at pH 3.75 was added to the above sample; MACS nano-magnetic beads (product of Miltenyi Biotechnology Co., Ltd., Germany) were added to the serum, and incubated on a magnet for 10-30 min; the supernatant was removed; Then add the binding reagent, place it on the magnet and incubate for 1-5 minutes to make the nano-magnetic...
Embodiment 2
[0057] The diagnosis of embodiment 2 gastric cancer
[0058] 1. Establish a standard fingerprint library of normal serum:
[0059] 1,000 healthy volunteers with three consecutive years of physical examination, half male and half male, aged 20 to 50, no family medical history, at least two generations of cancer-free patients, a few drops of blood were drawn, and treated according to the method of Example 2. The difference is: the pH of the binding reagent is 3.95.
[0060] 2. Establish a serum fingerprint database for gastric cancer;
[0061] 52 patients were diagnosed with gastric cancer by CT, MRI and other examinations, half male and half male, aged 40 to 60, all patients underwent surgical resection of the tumor, and the pathological results proved to be gastric cancer. After the whole blood was collected before the operation, it was processed according to the method under the sample collection item, and the serum was collected and stored at -80°C; it was processed accord...
Embodiment 3
[0065] Example 3 Diagnosis of rectal cancer by body fluid
[0066] 1. Establish a standard fingerprint library of normal urine
[0067] A total of 1,000 healthy volunteers with three consecutive years of physical examination, half male and half male, aged 20 to 50, no family medical history, no cancer patients for at least two generations. The collected urine was processed and analyzed by mass spectrometry according to the steps in Example 2, except that the pH of the binding reagent used was 3.55.
[0068] 2. Establish a urine fingerprint database for rectal cancer;
[0069] CT, MRI and other examinations confirmed rectal cancer in 278 patients, half male and half male, aged 40 to 60. All patients underwent surgical resection of the tumor, and the pathological results proved to be rectal cancer. The collected urine was processed and analyzed by mass spectrometry according to the steps in Example 1, except that the pH of the binding reagent used was 3.55.
[0070] 3. Compar...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com