Method for calculating and identifying protein-RNA interaction sites
A technology of interaction site and recognition method, which is applied in the field of biomacromolecule interaction recognition, can solve the problems of low prediction accuracy and inability to obtain high sensitivity and specificity at the same time, achieve good generalization performance and ensure prediction Ability, the effect of preventing overfitting
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
[0012] The following is a detailed description of the use of the method of the present invention for protein-RNA interaction site identification as an example, including the following steps:
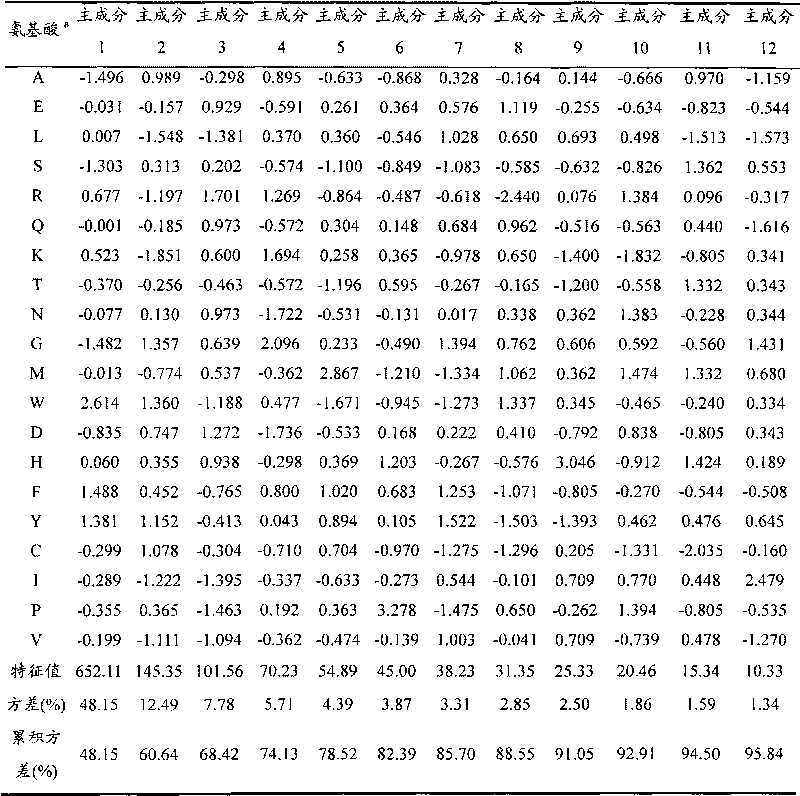
[0013] a) Select 640 kinds of two-dimensional property parameters of 20 kinds of natural amino acids, including: molecular electrical interaction vector, molecular electric distance vector and holographic molecular electric distance vector, topology, topological charge index, number of rotations and paths, edge adjacency index, Burden eigenvalue, autocorrelation, connectivity index, information index and eigenvalue index.
[0014] Table 1 12 principal component scores of 640 two-dimensional property parameters of 20 natural amino acids
[0015]
[0016] a The 20 natural amino acids are represented by conventional single English letters.
[0017] Using principal component analysis to process 640 kinds of property parameters, 12 principal components are obtained, which accumulatively ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com