Methicillin-resistant staphylococcus aureus (MRSA) recombinant multivalent subunit genetic engineering vaccine and method for preparing same
A genetically engineered vaccine, methicillin-resistant technology, applied in the field of biopharmaceuticals
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
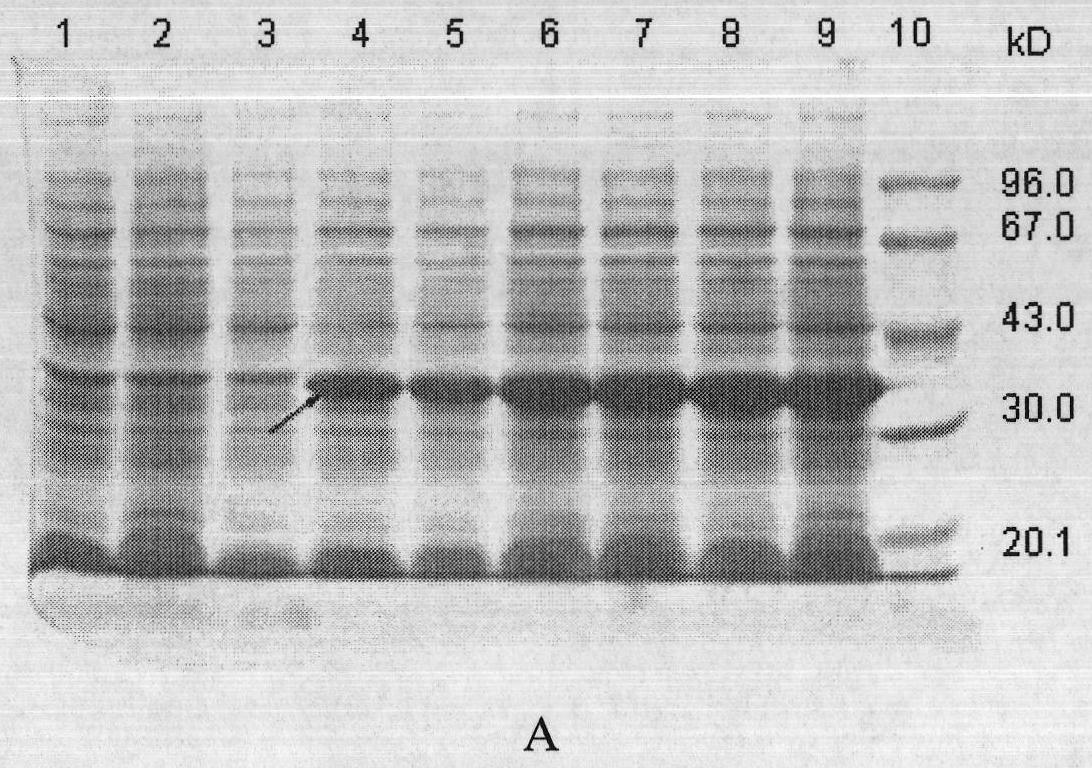
Examples
Embodiment 1
[0091] Cloning of embodiment 1 methicillin-resistant Staphylococcus aureus ClfA, IsdB, mSEC gene
[0092] 1. Methicillin-resistant Staphylococcus aureus WHO-2 (preserved by the Department of Pharmacology, Third Military Medical University)
[0093] 2. Take out the stored methicillin-resistant Staphylococcus aureus WHO-2 strain from the liquid nitrogen tank, spread it on the WHO-2 special solid medium, and cultivate it overnight at 37°C. The Genome Extraction Kit extracts the MRSA genome.
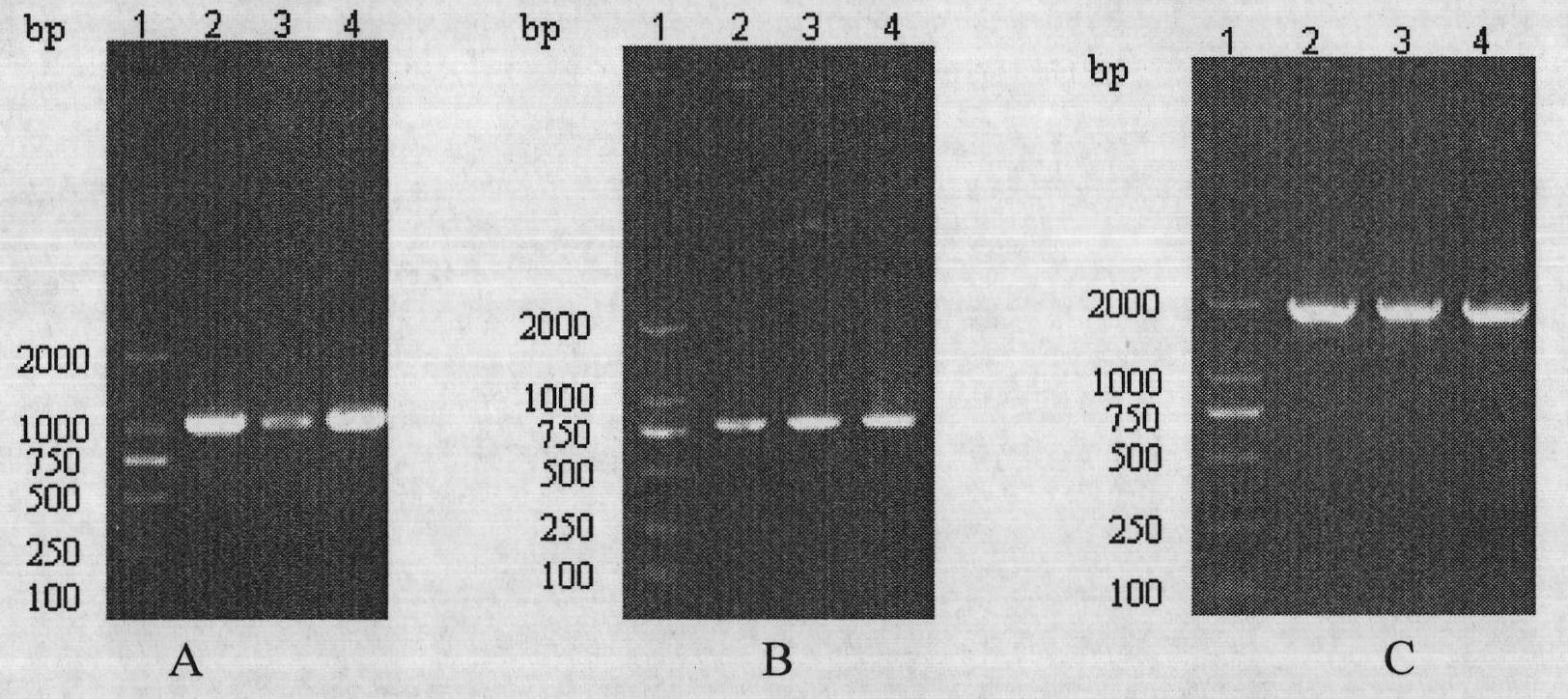
[0094] 3. The coding genes of CIfA, IsdB and mSEC were respectively amplified from the MRSA genome by PCR method.
[0095] 1) The primers were designed and synthesized as follows (the base sequence of the restriction site is underlined, and the base sequence of the linker is shown in gray)
[0096] According to the gene sequence published by GenBank and the principles of primer design, the corresponding primers were designed and restriction sites were introduced.
[0097] ClfA P1 5′TCG G...
Embodiment 2
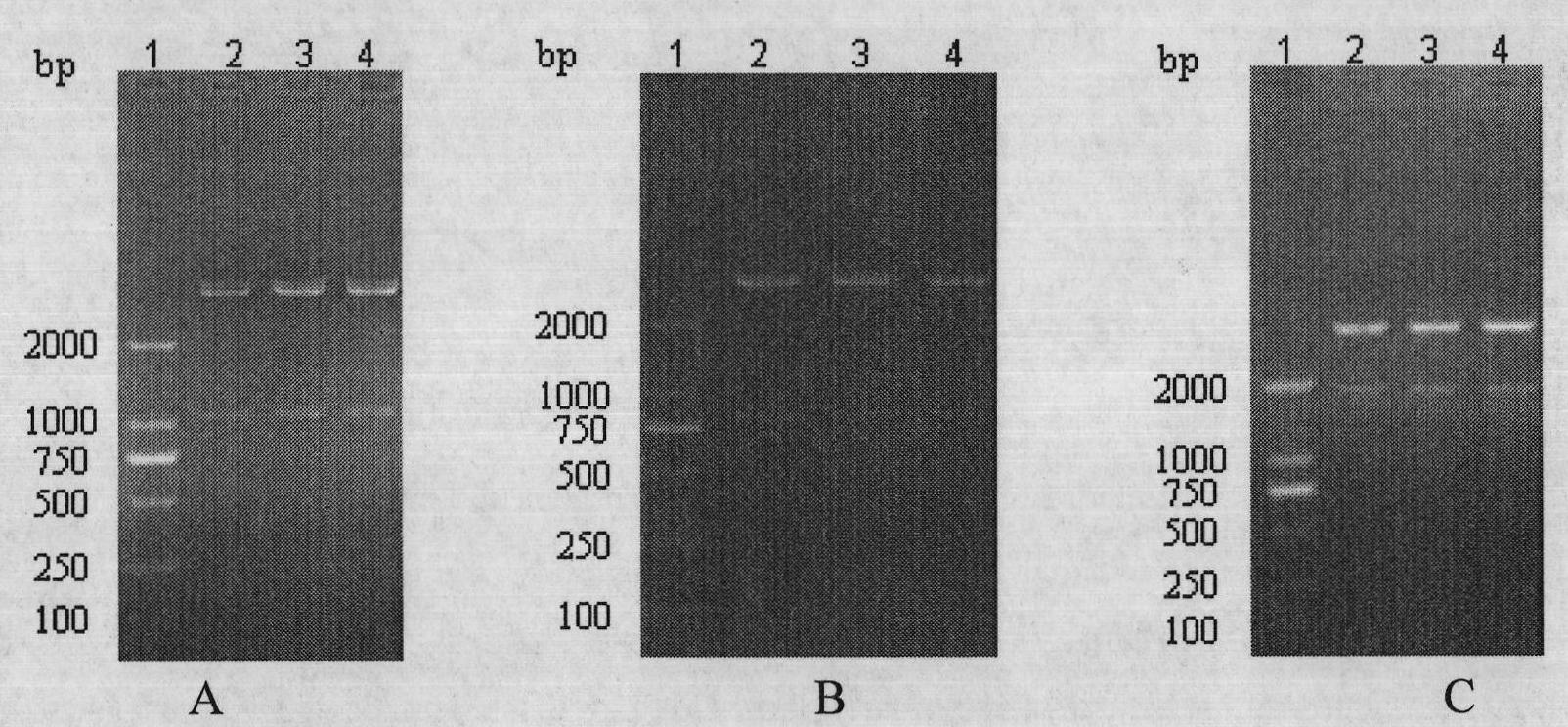
[0175] Example 2 ClfA 484-559 Fusion gene ClfA with mSEC 484-559 --Design and synthesis of primers for mSEC(CS)
[0176] ClfA 484-559 P1 5'
[0177] -TCG GGATCC ATGACAACACCATATATTGTAGTTGTTA-3′
[0178] Bam H I
[0179] P2 5′-TGAACCGCCTCCACCCCTCTGGAATTGGTTC-3′
[0180] mSEC P3 5′-GGTGGAGGCGGTTCAATGAAGTTATTTGCT-3′
[0181] P4 5′-TTTTTTGGTTAAATGAACTTCTACATTA-3′
[0182] Using the ClfA and mSEC recovered in Example 1 as templates, respectively amplify ClfA with P1 and P2, P3 and P4 primers 484-559 and mSEC gene, the PCR amplification system is: template DNA 1μl; 10×PCR buffer 5μl; dNTPs (10mmol / L) 4μl; primers (0.025mmol / L) 1μl; Taq DNA polymerase (5u / μl) 0.5μl ; Add deionized water to a final volume of 50 μl.
[0183] The reaction system was mixed evenly, and after centrifugation, 30 μl of paraffin oil was added. Pre-denaturation at 94°C for 5 minutes, denaturation at 94°C for 30 seconds...
Embodiment 3
[0186] Example 3 CS and IsdB 337-462 fusion gene CS-IsdB 337-462 (CSI) Acquisition
[0187] Primer design and synthesis
[0188] Add P1 5′-TCG to CS GGATCC ATGACAACACCATATATTGTAGTTGTTA-3′
[0189] Linker Bam H I
[0190] P5 5′-TGAACCGCCTCCACCTTTTTTGGTTAAATG-3′
[0191] IsdB 337-462 P6 5′-GGTGGAGGCGGTTCACCAACAAATGAAAAAATG-3′
[0192] P7 5′-GCG CTCGAG AGATTTATCGGTATTGGCTTTTGTA-3′
[0193] wxya
[0194] Respectively using the IsdB recovered in Example 1 and the CS recovered in Example 2 as templates, using P6 and P7, P1 and P5 as primers to amplify the IsdB with Linker respectively 337-462 and CS genes. The PCR amplification system is: template DNA 1μl; 10×PCR buffer (containing magnesium chloride) 5μl; dNTPs (10mmol / L) 4μl; primers (0.025mmol / L) 1μl; Taq DNA polymerase (5u / μl) 0.5μl , add deionized water to a final volume of 50 μl.
[0195] The reaction system was mixed evenly, and after centrifugation, 30 μl ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com