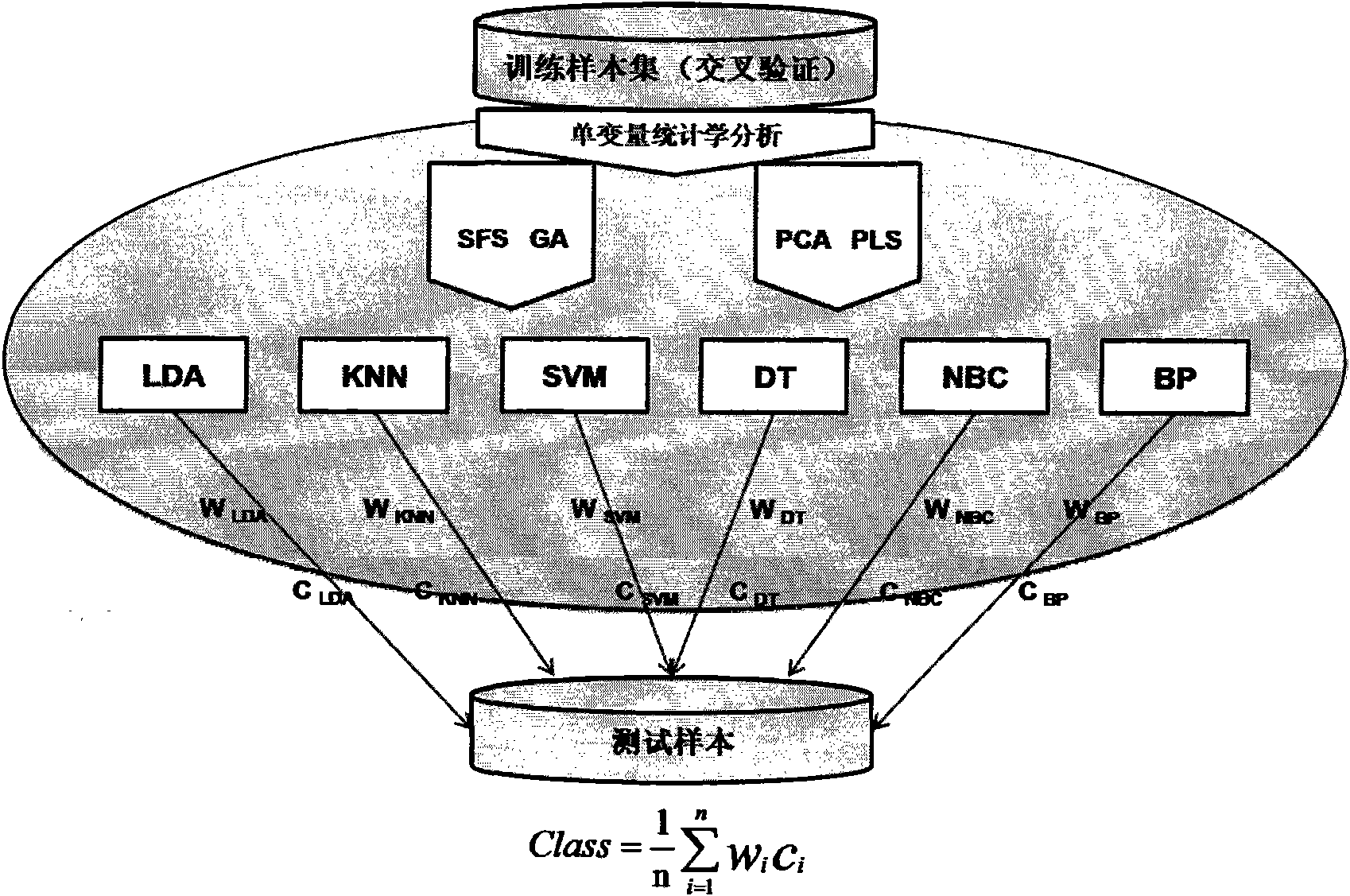
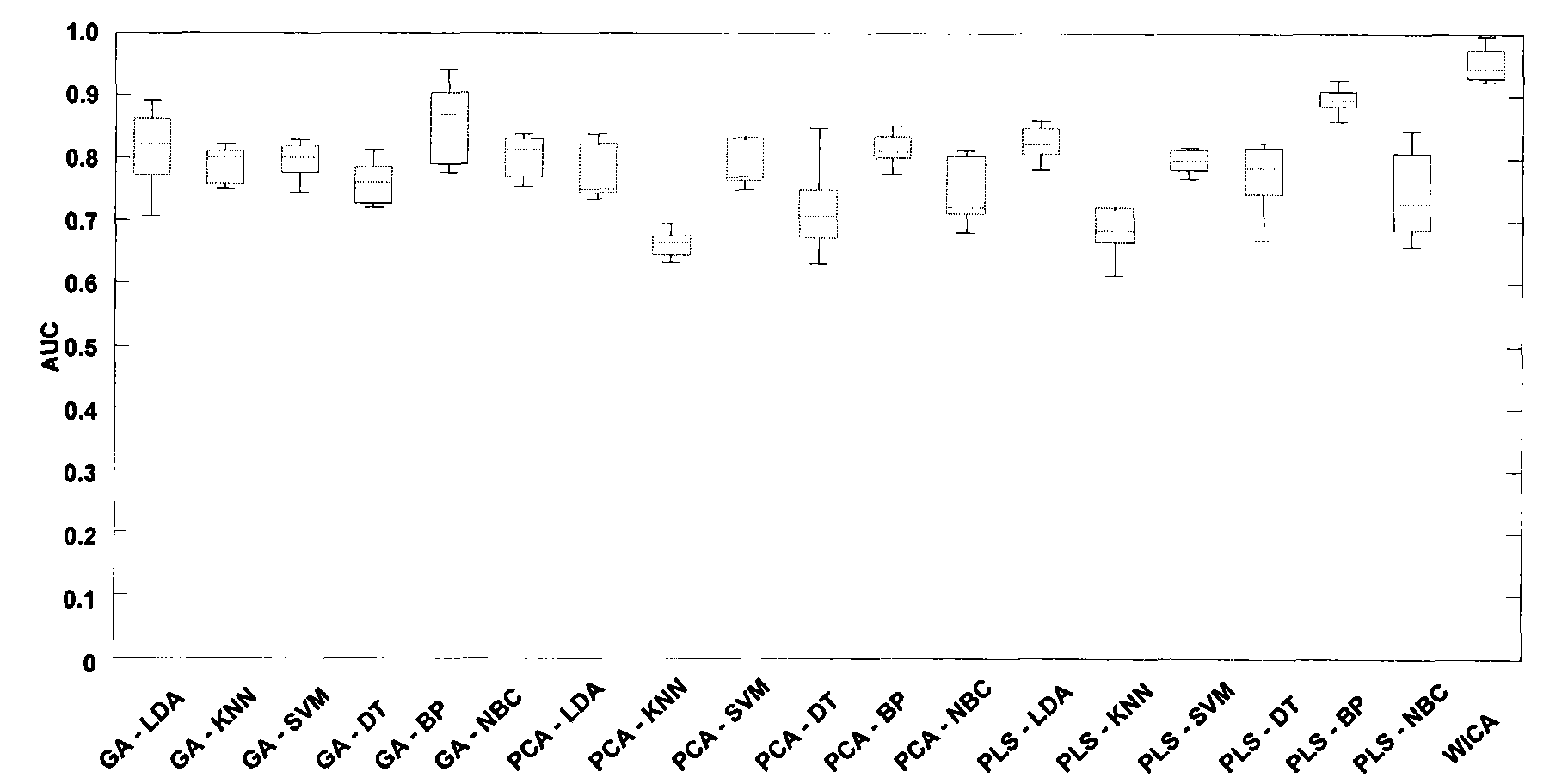
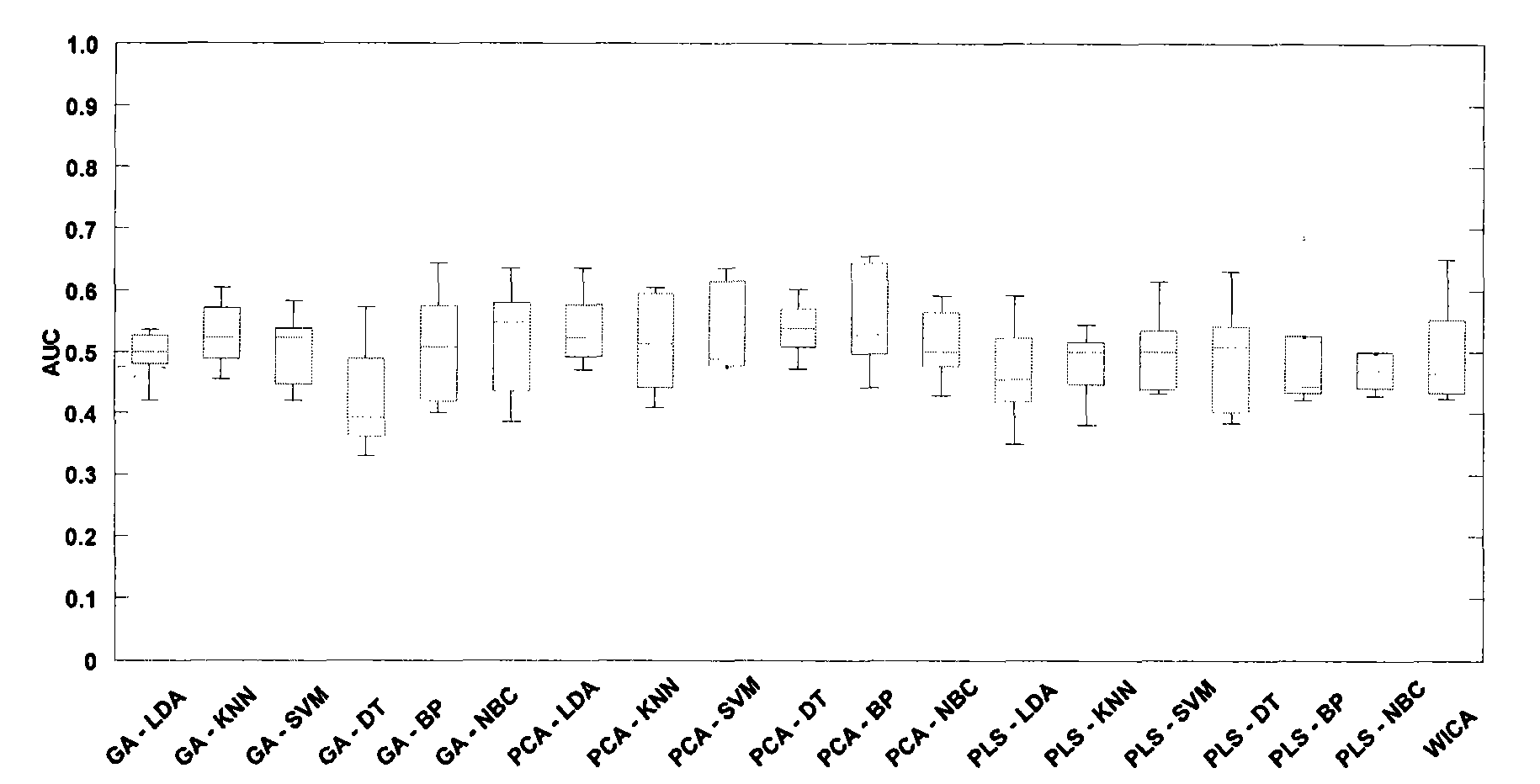Classification method of differential proteomics
A technology of proteomics and classification methods, applied in special data processing applications, instruments, electrical digital data processing, etc., to achieve high classification accuracy and robustness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] 1) Using two sets of differential proteomics public datasets widely used in the world as research materials
[0032] The first set of samples came from the National Cancer Institute (NCI). The data were divided into ovarian cancer samples and normal samples. The data were generated by the SELDI-TOF-MS analysis method, including 162 ovarian cancer samples and 91 normal samples. Dataset address: http: / / home.ccr.cancer.gov / ncifdaproteomics / ppatterns.asp. The second group of samples comes from the Keck laboratory of Yale University in the United States, and is divided into 93 ovarian cancer samples and 77 normal samples, which are produced by Micromass MALDI-L / R. In the present invention, the linear mode (Linear Mode) m / Z value is selected at Data sets ranging from 3450 to 28000Da were analyzed. Dataset address: http: / / bioinformatics.med.yale.edu / MSDATA2. In order to observe the effect of classifying the noise data (control) that is randomly grouped into samples, the clas...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com