DNA molecular label for high-throughput detection of human papilloma virus
A high-throughput, labeling technology, applied in the field of human papillomavirus detection, can solve the problems of high cost, low throughput, time-consuming and labor-intensive, and achieve the effect of low-cost detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0101] Example 1: Sample Extraction
[0102] According to the manufacturer's instructions, DNA was extracted from 190 exfoliated cells with known HC-II results using a KingFisher automatic extractor (ThermoScientific Kingfisher Flex automatic magnetic bead extraction and purification system, USA). Nucleic acid extraction was performed using the program "Bioeasy-200 μl BloodDNA-KF.msz". After the program is finished, about 100 μl of the eluted product (extracted DNA) is obtained, which is used as a template in the next step of PCR amplification.
Embodiment 2
[0103] Embodiment 2: PCR amplification
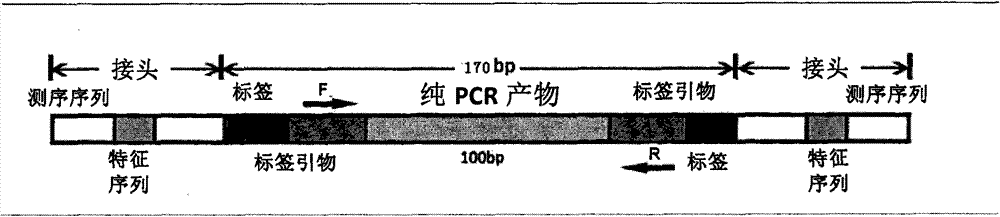
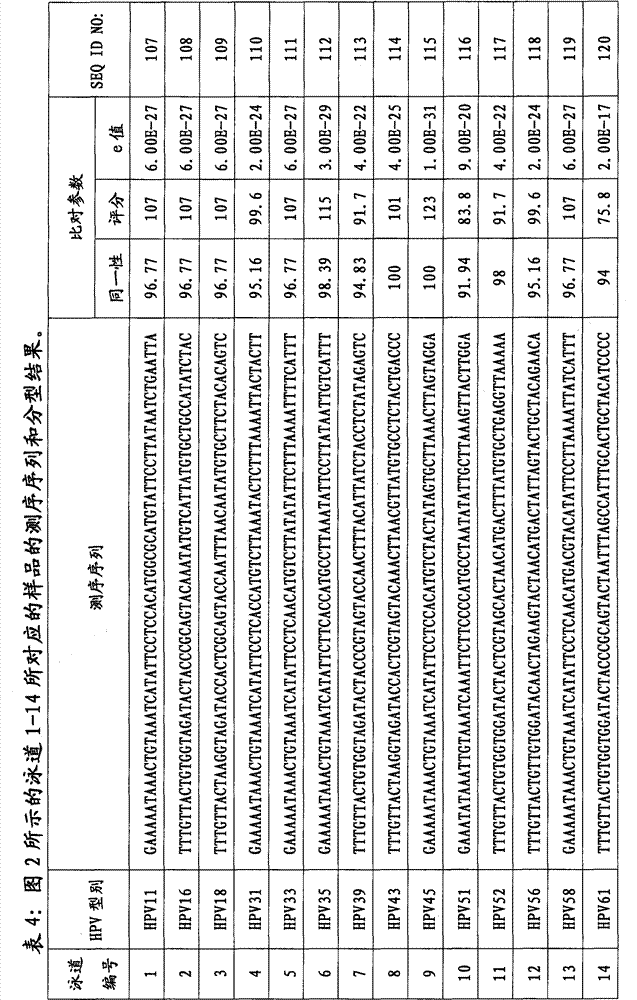
[0104] The 190 copies of DNA obtained in Example 1 were numbered 1-190 in sequence, and were evenly divided into 2 groups (HPV-1 group: numbering 1-95; HPV-2 group: numbering 96-190). According to the sequence (Table 2, SEQ ID NO:96-107) of each primer of the primer set (including 6 forward primers and 5 reverse primers) used to amplify HPV DNA, design a set of tags, a total of 95 (Table 1, SEQ ID NO: 1-95). Add each designed tag to the 5' end of the sequence of each primer in the primer set to obtain 95 tag primer sets, where each tag primer set includes corresponding 6 forward tag primers and 5 reverse Index primers, and different index primer sets use different tags (ie, 95 index primer sets correspond to 95 tags one-to-one).
[0105] PCR reactions were performed for all samples in 96-well plates, using a total of 2 plates (1 plate each for HPV-1 group and HPV-2 group). Using the DNA obtained in Example 1 as a template, and in t...
Embodiment 3
[0121] Example 3: Mixing and purification of PCR products
[0122] Mix the remaining PCR products of the HPV-1 group and the HPV-2 group in a 3ml EP tube (also marked as HPV-1 group and HPV-2 group), and shake to mix. 500 μl of DNA was taken from each of the 2 tubes of mixture, and purified by column using Qiagen DNA Purification kit according to the manufacturer's instructions to obtain 200 μl of DNA. Using Nanodrop 8000 (Thermo Fisher Scientific), the DNA concentrations of the purified mixtures were determined to be 98 ng / μl (HPV-1 group) and 102 ng / μl (HPV-2 group), respectively.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com