siRNA that inhibits porcine somatostatin receptor 2 gene expression
A somatostatin receptor and gene expression technology, which is applied in the fields of molecular biology and bioengineering, can solve problems such as unseen research reports, and achieve the effects of improving growth performance, good silencing effect, and promoting growth
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
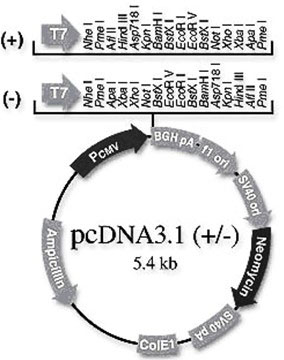
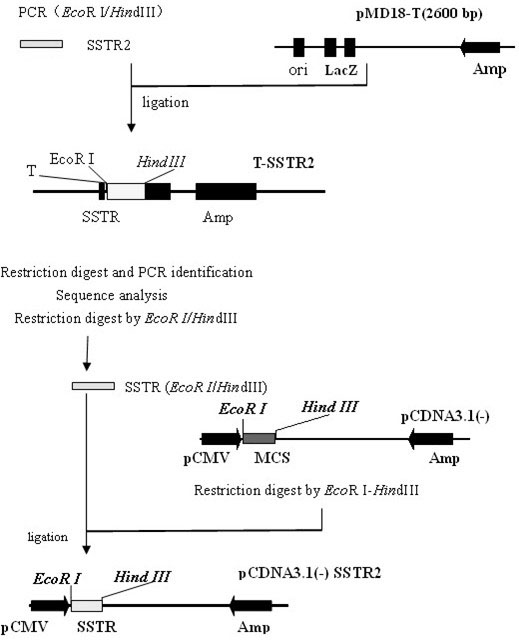
[0040] Example 1 Construction of pCDNA3.1-SSTR2 eukaryotic expression vector
[0041] 1. The first strand of cDNA was synthesized using the normal porcine pituitary total RNA preserved in our laboratory as a template;
[0042] 2. According to the sequence of porcine somatostatin receptor 2 (SSTR2) (GenBank accession number NM001011694), design a pair of primers and introduce restriction restriction sites Eco R I and Hin d III, the sequence of the upstream primer is SEQ ID NO:10, and the sequence of the downstream primer is SEQ ID NO:11. Using the first strand of cDNA as a template, the SSTR2 gene was amplified by PCR. Reaction system: 5 μL of 10×Ex Taq Buffer, 4 μL of 2.5mM dNTP, 1 μL of upstream primer (10 pmol / μL), 1 μL of downstream primer (10 pmol / μL), 3 μL of cDNA template, 1 μL of Ex Taq DNA polymerase (5 U / μL), supplement ddH 2 0 to 50 μL. Reaction conditions: 94°C, 5min; 94°C, 30sec; 56°C, 30sec; 72°C, 1min, a total of 35 cycles; 72°C, 10min extension and then...
Embodiment 2
[0050] Example 2 Construction of pshRNA-copGFP Lentivector lentiviral interference plasmid
[0051] 1. According to the method and principle of porcine somatostatin receptor 2 mRNA sequence design, target sequences were screened out, and siRNA was designed to be reverse complementary to the mRNA of somatostatin receptor 2 gene, named SSTR2-1, SSTR2-2, and SSTR2 respectively -3, its sense strand sequence is as follows:
[0052] SSTR2-1: SEQ ID NO: 1;
[0053] SSTR2-2: SEQ ID NO: 2;
[0054] SSTR2-3: SEQ ID NO:3.
[0055] 2. According to the siRNA target sequence, design the DNA sequence required to construct the siRNA vector that inhibits the expression of the porcine somatostatin receptor 2 gene: SSTR2 shRNA template, introduced at both ends Bam H I and Eco The R I enzyme cutting site was sent to Shanghai Bioengineering Technology Service Co., Ltd. for synthesis. The specific sequence is as follows (F indicates the sense strand, R indicates the antisense strand):
[...
Embodiment 3
[0068] Example 3 Screening of Effective Targets of RNA Interference
[0069] 1. Transfection of eukaryotic cells with three lentiviral interference plasmids
[0070] pcDNA3.1-SSTR2 and LV-siRNA1, LV-siRNA2 and LV-siRNA3 were co-transfected into CHO cells for expression. CHO cells were co-transfected with pcDNA3.1-SSTR2 and LV-shRNA-GFP empty plasmids as a control, in order to be consistent with the transfection background of the treatment group, and three replicates were set for each treatment. The ratio of pcDNA3.1-SSTR2 to lentiviral interference plasmid is 1:5.
[0071] For lipofection, follow Lipofectamine TM 2000 Reagent Manual.
[0072] (1) One day before transfection, seed the cells in a new culture dish and blow the cells completely to ensure the uniform distribution of the cells. When transfecting, the cells should ideally cover 60-70% of the dish.
[0073] (2) Add a total of 4 μg of pcDNA3.1-SSTTR2 and lentiviral interference plasmid at a ratio of 1:5 to 250 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com