Method for rapid detection of Listeria monocytogenes viable bacteria in liquid milk
A technology of Listeria monocytogenes and liquid milk, which is applied in the field of detection of viable Listeria monocytogenes, can solve the problems of high detection cost, long detection time, inability to effectively distinguish dead bacteria and live bacteria, etc., and achieve high detection sensitivity , time-consuming, shorten the effect of detection time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
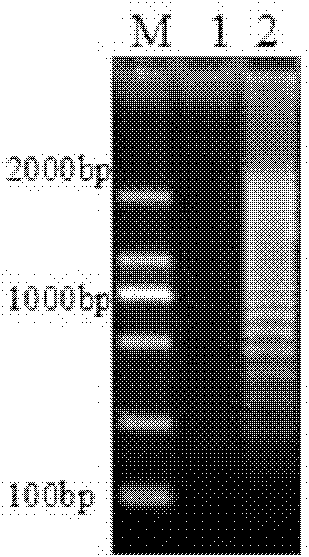
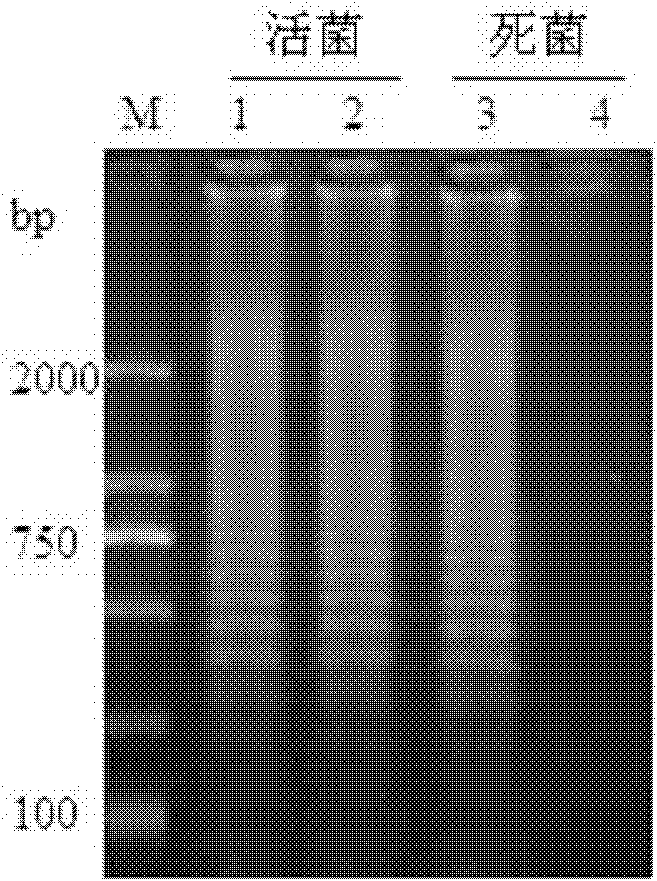
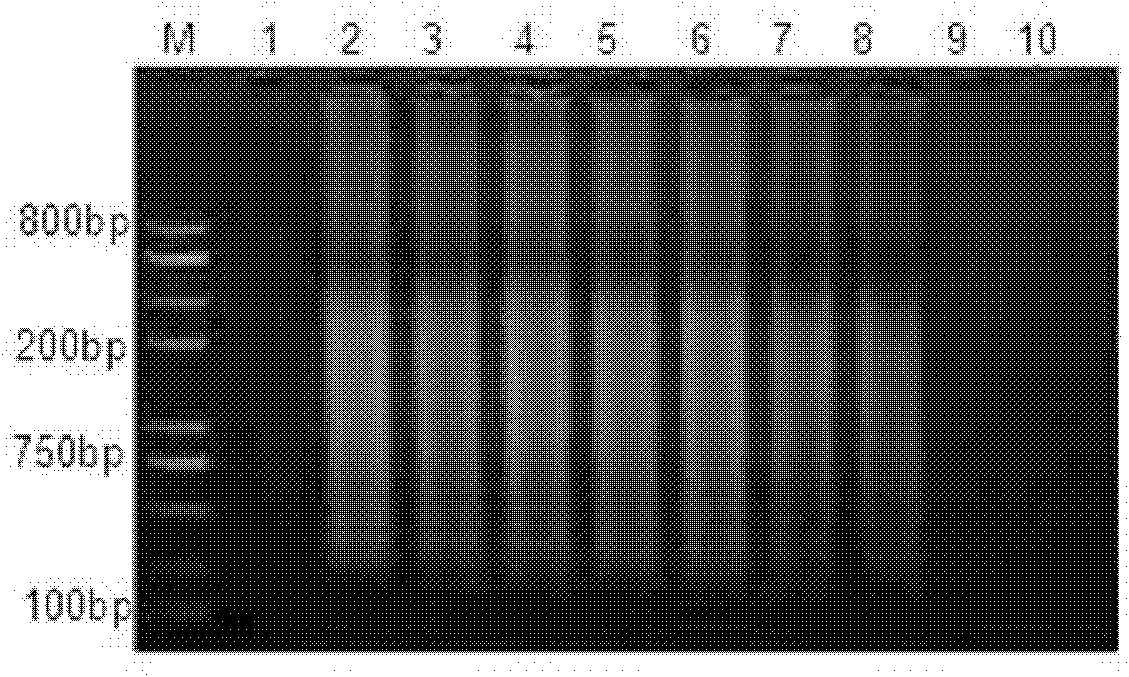
[0011] Specific embodiment one: the method for quickly detecting the live bacteria of Listeria monocytogenes in liquid milk in this embodiment is carried out according to the following steps: 1. Pretreatment of the tested milk sample: add PMA to 1mL of the tested milk to make the PMA The concentration is 50μmol / L, then placed in a dark place for 5-30min, then placed 15-20cm away from a 650W halogen lamp, and exposed for 5-30min; Add 150 μL of sodium citrate with a concentration of 250 g / L and 100 μL of sodium hydroxide with a concentration of 1 mol / L, then centrifuge at 10,000 r / min for 2 minutes, discard the supernatant, and obtain a precipitate; 3. Wash with 0.85% normal saline Precipitate, then centrifuge at 10000r / min for 1min, discard the supernatant, and use the bacterial genome DNA extraction kit to extract DNA to obtain the extract; 4. Use the extract as a template for LAMP amplification to obtain the LAMP amplification product; 5. Identification : Take 5 μL of LAMP am...
specific Embodiment approach 2
[0014] Embodiment 2: This embodiment is different from Embodiment 1 in that: in step 1, place it in a dark place for 10-20 minutes. Others are the same as in the first embodiment.
specific Embodiment approach 3
[0015] Embodiment 3: This embodiment is different from Embodiment 1 in that: in step 1, place it in a dark place for 15 minutes. Others are the same as in the first embodiment.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com