Method for screening protein aptamers by using micro-fluidic chip
A microfluidic chip and nucleic acid aptamer technology, which is applied in the field of screening target protein-specific nucleic acid aptamers using a specific device, can solve the problem of fewer types of nucleic acid aptamers, shorten the screening time, reduce the number of cycles, and improve the The effect of filtering strength
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
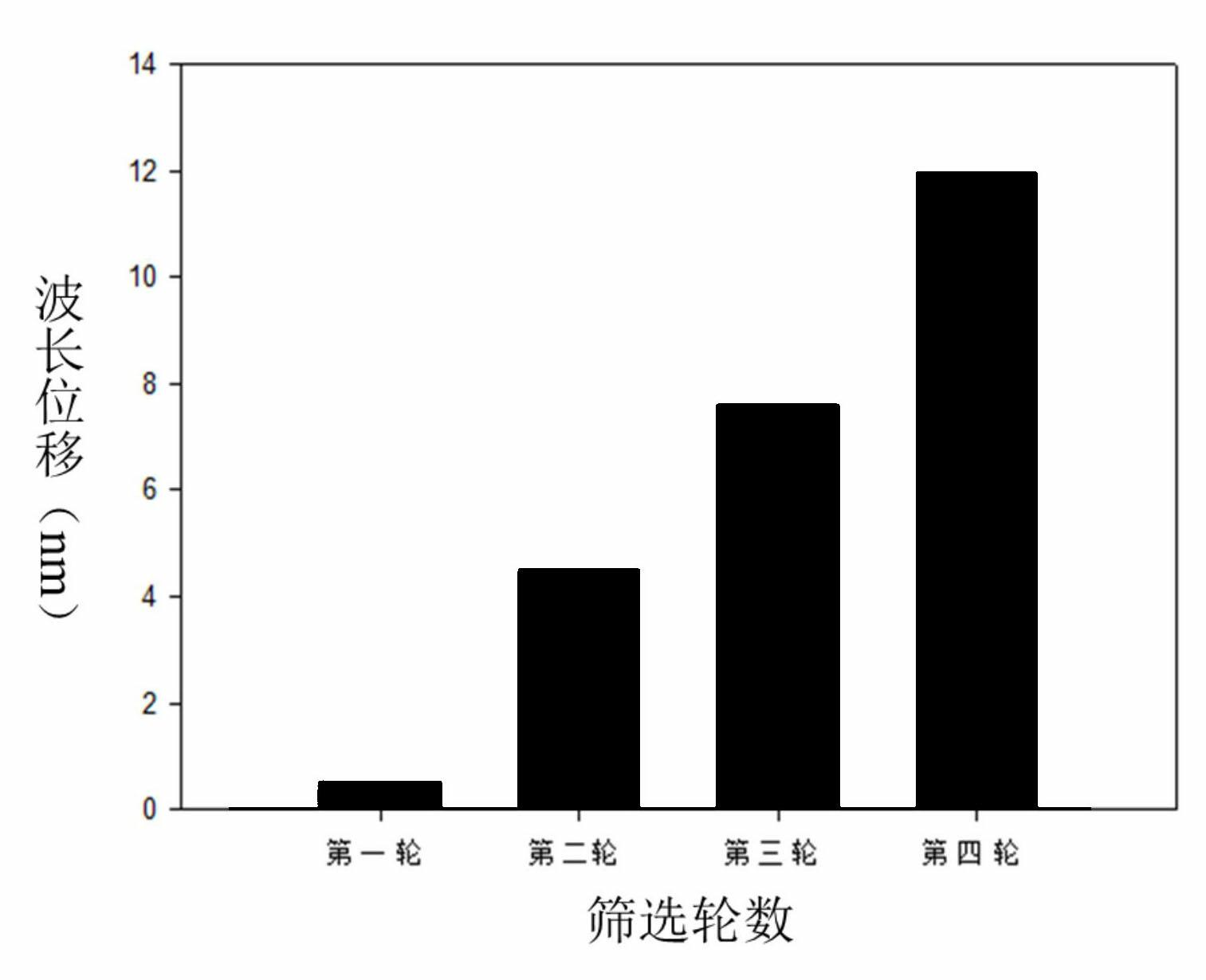
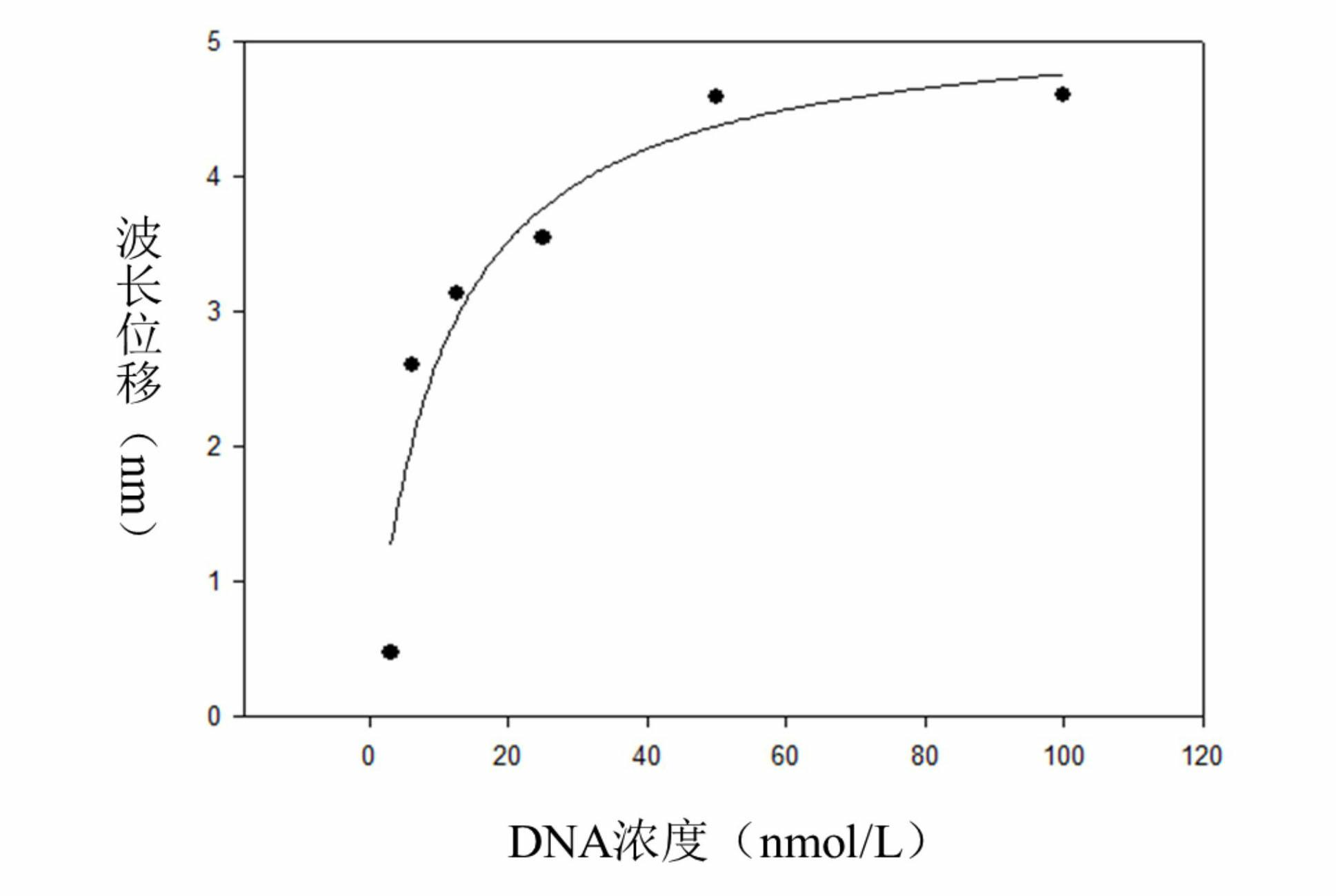
[0023] Example 1: Screening of nucleic acid aptamers specifically binding to myoglobin.
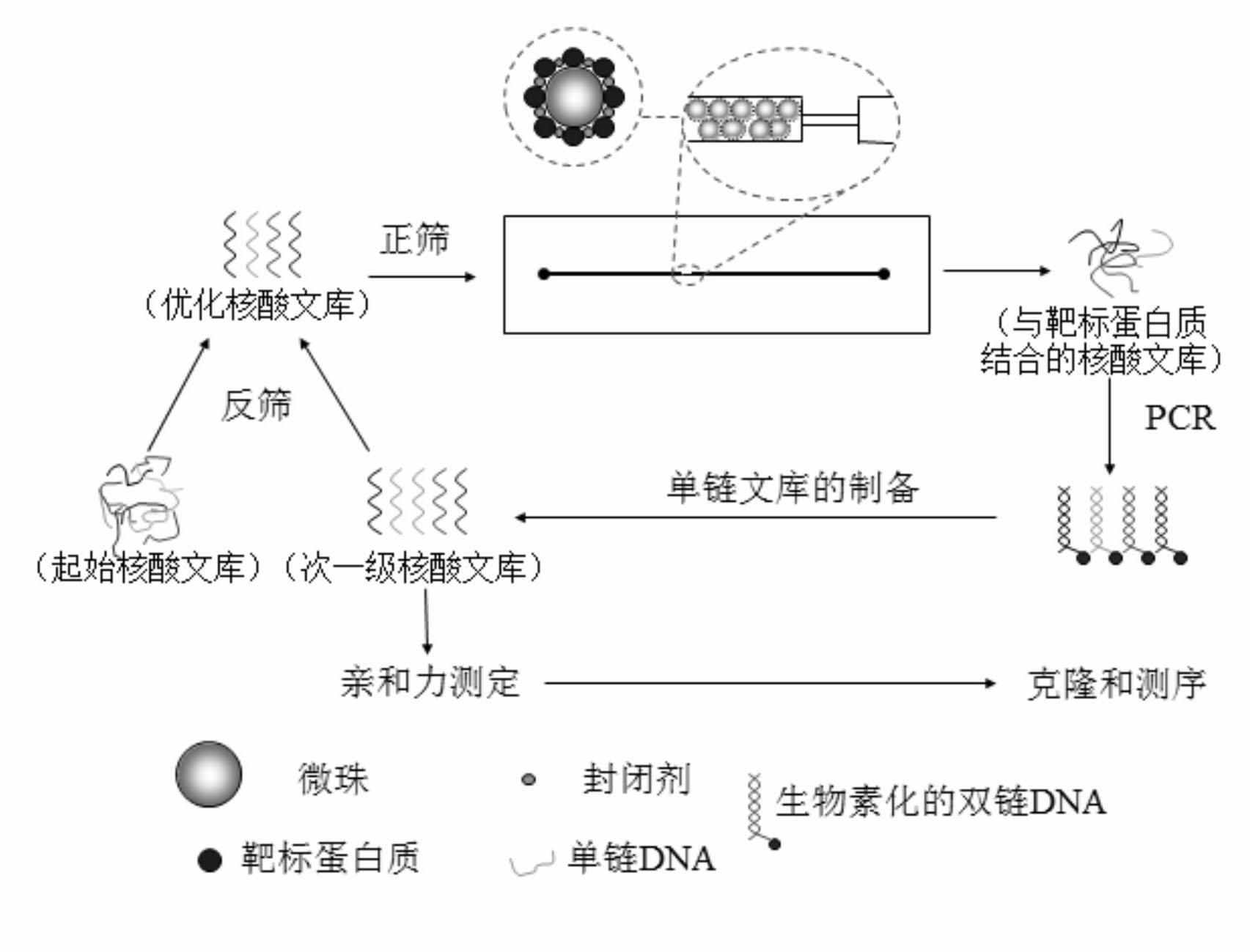
[0024] a kind of like figure 1 The method for utilizing microfluidic chip screening myoglobin nucleic acid aptamer of the present invention as shown, comprises the following steps:
[0025] 1. Optimizing Nucleic Acid Libraries
[0026] 1.1 Design the initial nucleic acid library: the capacity of the initial nucleic acid library is 10 14 Above, design and synthesize a random nucleic acid sequence library comprising 20 nucleotides at both ends and 40 nucleotides in the middle as follows:
[0027] 5’-CCGTTGCTACCGAGTGTCTG-N 40 -ATGAAGAAAGAGGAACGGGC-3'.
[0028] 1.2 Optimizing the starting nucleic acid library: dissolve the synthesized 200pmol single-stranded starting nucleic acid library in binding buffer (20mmol / L Hepes, 120mmol / L NaCl, 5mmol / L KCl, 1mmol / L CaCl 2 , 1mmol / L MgCl 2 , pH 7.35), heat denaturation treatment in it, heat at 95°C for 5min, place on ice for 10min, and then p...
Embodiment 2
[0048] Example 2: Screening of nucleic acid aptamers specifically binding to C-reactive protein.
[0049] a kind of like figure 1 The method for utilizing microfluidic chip screening myoglobin nucleic acid aptamer of the present invention as shown, comprises the following steps:
[0050] 1. Optimizing Nucleic Acid Libraries
[0051] 1.1 Design the initial nucleic acid library: the capacity of the initial nucleic acid library is 10 14 Above, design and synthesize a random nucleic acid sequence library comprising 16 nucleotides at both ends and 40 nucleotides in the middle as follows:
[0052] 5’-GGCAGGAAGACAACA-N 40 -TGGTCTGTGGTGCTGT-3'.
[0053] 1.2 Optimizing the starting nucleic acid library: dissolve the synthesized 200pmol single-stranded starting nucleic acid library in binding buffer (20mmol / L Hepes, 120mmol / L NaCl, 5mmol / L KCl, 1mmol / L CaCl 2 , 1mmol / L MgCl 2 , pH 7.35), heat denaturation treatment in it, heat at 95°C for 5min, place on ice for 10min, and then p...
PUM
| Property | Measurement | Unit |
|---|---|---|
| width | aaaaa | aaaaa |
| height | aaaaa | aaaaa |
| height | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com