Hyper-branched rolling cycle amplification detection method of soft-shelled turtle iridovirus
A technology of iridescent virus and rolling circle amplification, applied in the direction of microorganism-based methods, microorganism measurement/inspection, biochemical equipment and methods, etc., can solve the problems of inability to detect diseases in time and accurately, increase the risk of cross-infection, and detection sensitivity Low-level problems, to achieve the effect of increasing the chance of cross-contamination, reducing the risk of reaction contamination, and high sequence specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] 1. Preparation of DNA sample template and standard sample of SITV genome
[0032]1.1 Extraction of Chinese soft-shelled turtle iridescent virus DNA
[0033] 1.1.1 Purification of virus
[0034] Cut 20 grams of infected tissue into small pieces, put them in 200 ml of TNE buffer, centrifuge at 8000 r / min, 4°C for 20 min, and take the supernatant. Centrifuge at 35000g for 1h at 4°C. Discard the supernatant, except for the precipitated black circle and other precipitates. Resuspend in 5ml TM buffer. Overnight, gently pipette the overnight precipitate evenly, centrifuge at 8000r / min, 4°C for 20min to take the supernatant, and centrifuge at 35000g for 1h at 4°C, the obtained white precipitate is the purified virus.
[0035] 1.1.2 Extraction of purified viral DNA
[0036] Completely transfer the virus to a centrifuge tube with 500 µl of TE buffer. Add 125 microliters of 10% SDS to the tube to make a final concentration of 2%. Add proteinase K to the EP tube to make a fi...
Embodiment 2
[0057] The present invention detects the sensitivity of the STIV method based on HRCA technology
[0058] 1. Preparation of standard substance: measure the concentration of the extracted DNA sample with a spectrophotometer and dilute to 10 10 copies / μL as a standard. Dilute the prepared standard to 10 0 -10 5 copies / μL were used as templates respectively.
[0059] 2. Use the optimal condition method obtained in Example 1 to detect the templates of each concentration, thereby obtaining the sensitivity of the present invention, and perform conventional PCR detection in addition to compare the sensitivity.
[0060] 2.1 Connection of lock probe
[0061] The ligation reaction system is as follows: padlock probe 0.1μM, T4DNA Ligase 1U / μl, 10×T4DNA Ligase Buffer 1μl, sample template DNA 2μl, add water to a total reaction volume of 10μl, and the ligation conditions are: after denaturation at 94°C for 5 minutes, Add 1U / μl T4 DNA Ligase, and react for 10 at 16°C, and the ligation r...
Embodiment 3
[0070] The present invention is based on the specificity determination of the method for detecting STIV by rolling circle amplification technology
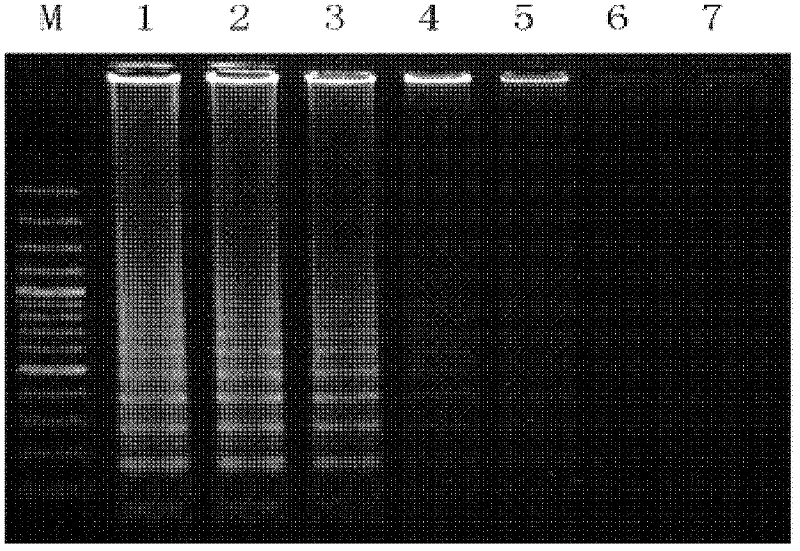
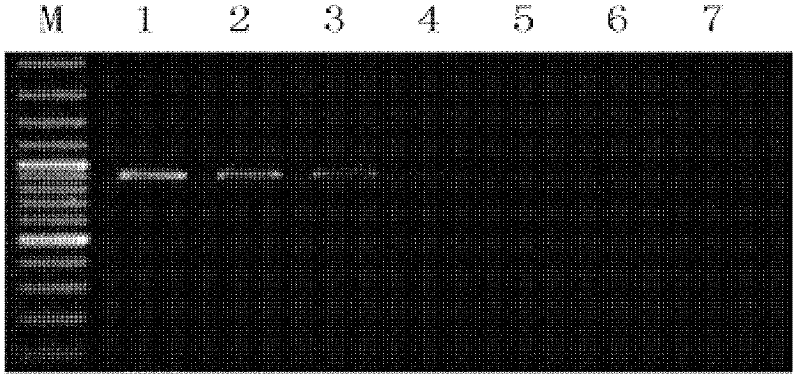
[0071] STIV genomic DNA was used as a positive control, and three other viruses with different degrees of correlation were used to verify the specificity of this study, namely white spot syndrome virus (WSSV), Singapore grouper iridovirus (Singapore grouperiridovirus, SGIV) and Portunus reovirus, wherein double distilled water was used as a negative control. Amplified products were detected by agarose gel electrophoresis. The results show that see image 3 , indicating that the HRCA detection method provided by the present invention can ensure the specific detection of STIV and does not cross-react with other related viruses.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com