Improved spectrophotometry method for determining proteins by using Coomassie brilliant blue
A Coomassie brilliant blue method and a technology of Coomassie brilliant blue, which are applied in the field of photometry for the determination of proteins by Coomassie brilliant blue, can solve problems such as poor protein stability, and achieve the effect of improving stability and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Embodiment 1 (determination of protein in penicillin sodium salt)
[0025] Sample pretreatment: Accurately weigh 0.4800g of penicillin sodium for injection produced by a pharmaceutical company in a beaker, add appropriate amount of water to dissolve, transfer to a 50mL volumetric flask after dissolution, and shake to volume with water.
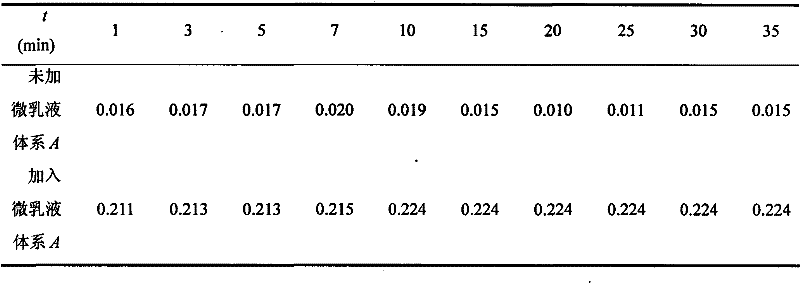
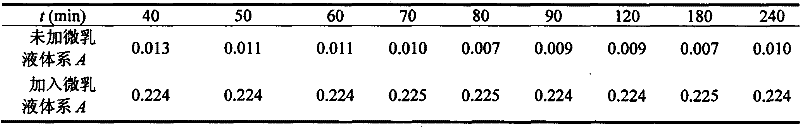
[0026] According to the mass ratio OP-10: n-butanol: n-heptane: water=2:3:1:90, prepare OP-10 microemulsion, add 2.00mL Coomassie brilliant blue G-250 solution (10.0 μg·mL -1 ), 4.00μLOP-10 microemulsion, 1.00mL sample solution, dilute to volume with water and shake well, let it stand for 25min, and measure the absorbance of the system at 595nm with the reagent blank as a reference. The measurement results are shown in Table 3.
[0027] The analysis result of protein in the penicillin sodium salt of table 3
[0028]
Embodiment 2
[0029] Embodiment 2 (determination of protein in penicillin G potassium industrial salt)
[0030] Sample treatment: Accurately weigh 0.9600g of penicillin G potassium industrial salt produced by a certain company in a beaker, add appropriate amount of water to dissolve, transfer to a 100mL volumetric flask after dissolution, add water to volume and shake well.
[0031] According to the mass ratio OP-10: n-butanol: n-heptane: water=1:4:1:92, OP-10 microemulsion was prepared, and 2.00mL Coomassie Brilliant Blue G-250 solution (10.0 μg·mL -1 ), 4.00μL OP-10 microemulsion, 1.00mL sample solution, shake to a constant volume, let it stand for 15min, and measure the absorbance of the reagent blank at 595nm. The measurement results are shown in Table 4.
[0032] The analysis result of protein in the penicillin G potassium industrial salt sample of table 4
[0033]
Embodiment 3
[0034] Embodiment 3 (determination of protein in milk powder sample)
[0035] Sample treatment: Accurately weigh 0.0313g of commercially available milk powder into a glass mortar, dilute the sample solution to a 100mL volumetric flask, dilute to volume with water and shake well.
[0036] According to the mass ratio OP-10: n-butanol: n-heptane: water=2:3:1:89, prepare OP-10 microemulsion, add 2.00mL Coomassie brilliant blue G-250 solution (10.0 μg·mL-1), 4.00μL OP-10 microemulsion, 2.50mL sample solution, shake at constant volume, let stand for 20min, measure the absorbance of the reagent blank at 595nm. The measurement results are shown in Table 5.
[0037] The analysis result of protein in the milk powder sample of table 5
[0038]
[0039] As can be seen from the measurement results of the above 3 embodiments, the relative standard deviation is less than 5%, indicating that the method precision is higher; the recovery test shows that the recovery rate recorded is all wi...
PUM
| Property | Measurement | Unit |
|---|---|---|
| absorbance | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com