Affinity prediction and analysis method for human amphiphysin-1 Src homology-3 domain binding peptide
A homologous domain and analysis method technology, applied in special data processing applications, instruments, electrical digital data processing, etc., can solve the problems of unsuitability for high-throughput screening, low efficiency, time-consuming and labor-intensive, and achieve easy operation. , the effect of clear meaning and strong representational ability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
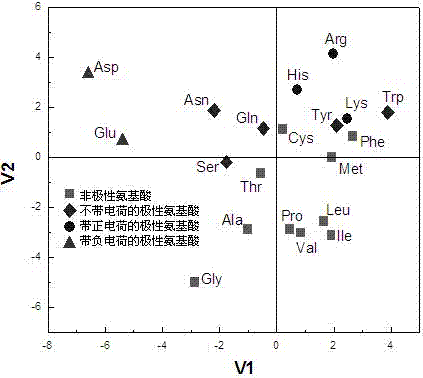
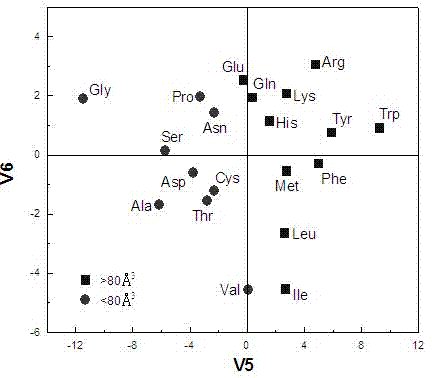
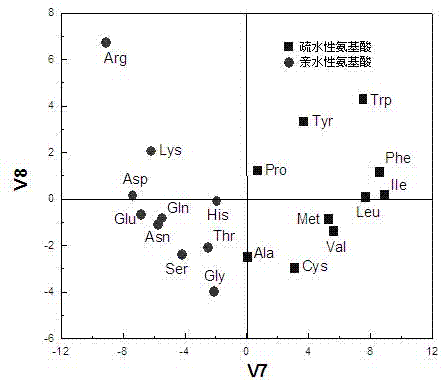
[0027] In order to make the purpose, technical scheme and advantages of the present invention more clear, the following will take the decapeptide as an example, in conjunction with the accompanying drawings, the affinity prediction and analysis method of the human type 1 biporin Src homology domain 3 binding peptide will be described in detail describe.
[0028] 1. Based on principal component analysis, the classification score of amino acid physicochemical properties was established
[0029]Non-bonding interactions play an important role in the mutual recognition and binding of peptides and proteins, mainly manifested as electronic, stereo, hydrophobic and hydrogen bond interactions, and other interactions such as charge transfer, salt bridge, etc. can be regarded as the special effects of the former. Manifestations. Considering that peptides and proteins are mainly combined through non-bond interactions, and amino acids are the basic structural modules of peptides and prote...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com