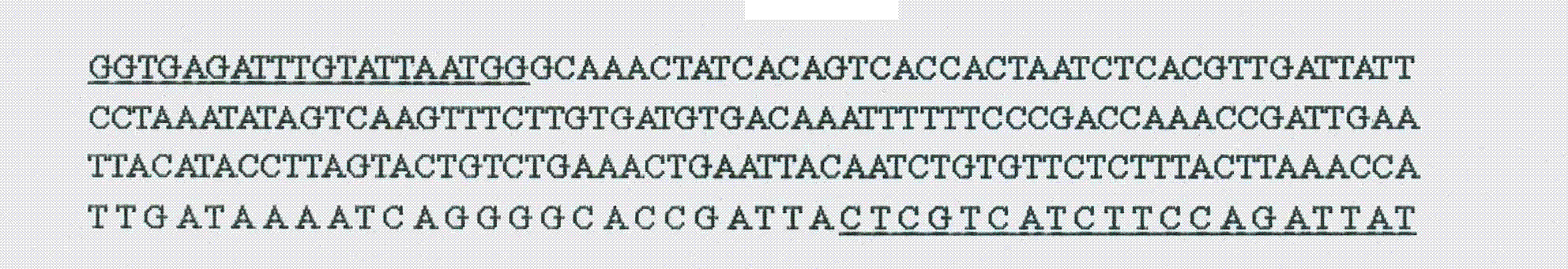Specific nucleic acid identification sequence used for detecting proteus mirabilis, and application thereof
A technology of proteus mirabilis and identification sequence, which is applied in the field of specific nucleic acid identification sequence to achieve the effect of short time, high sensitivity and specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] 2. Design and verification of the specific nucleic acid identification sequence of Proteus mirabilis
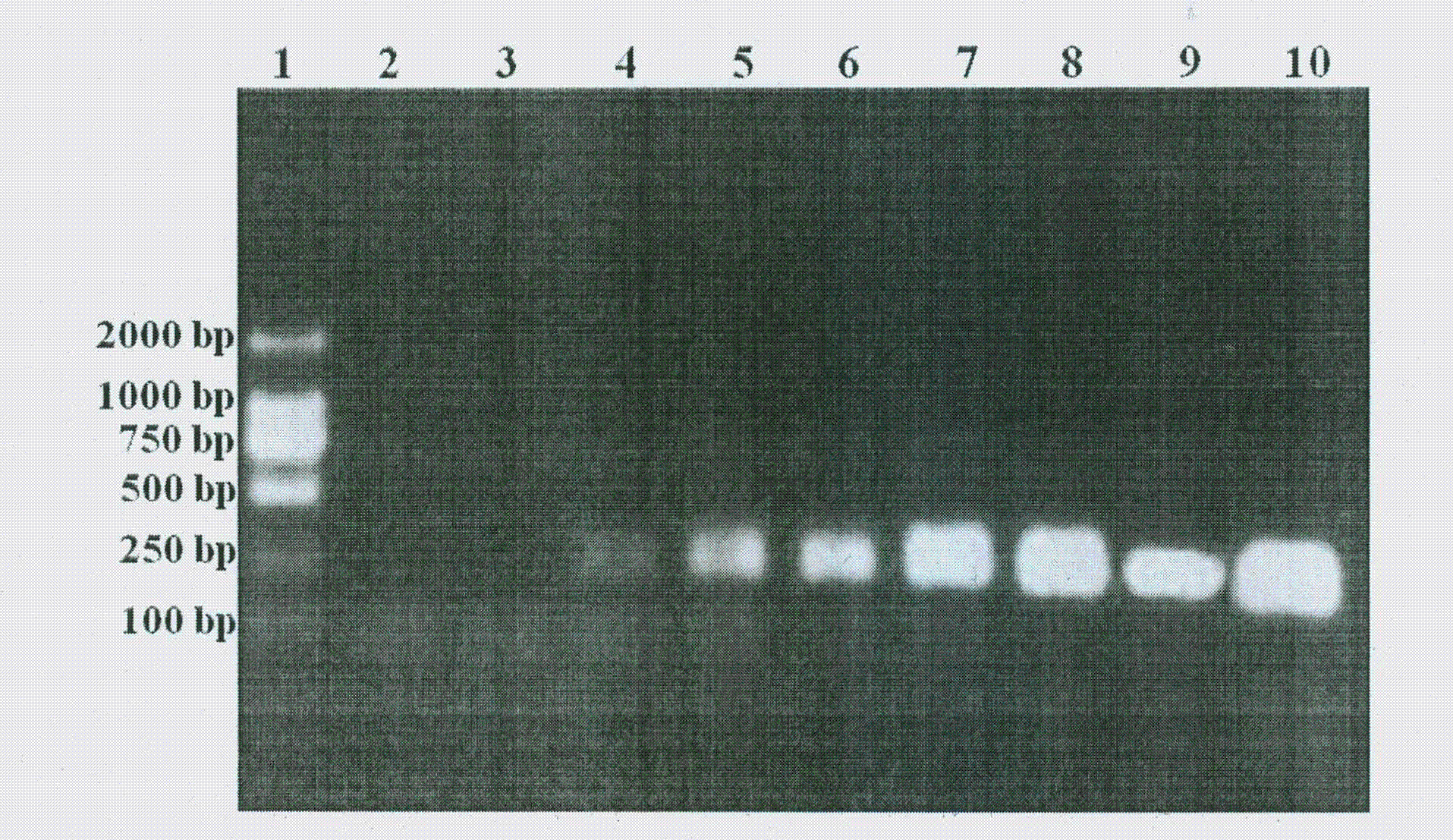
[0041] (1) According to the nucleic acid sequence of Proteus mirabilis ureR, primers ureRF1: 5'-GGTGAGATTTGTATTAATGG-3' and ureRR1: 5'-ATAATCTGGAAGATGACGAG-3' were designed. Use this nucleic acid identification sequence ureRF1 and ureRR1 as the forward and reverse primers of the PCR reaction respectively, and use the isolated Proteus mirabilis V7 (CGMCC No.4312) and Providencia stutzeri containing the ureR homologous gene, Escherichia Bacillus, Escherichia coli, and some common pathogens in the environment (strains include Vibrio, Edwardsiella, Klebsiella, and Pseudomonas strains) as template PCR to determine the use of PCR for the detection of Proteus mirabilis specificity.
[0042] (2) 15 μl of the PCR reaction system were added the following volumes of substances: 1.5 μl of 10×PCR buffer (200 mmol l -1 Tris-HCl (pH 8.4), 200mmol l -1 KCl, 15 mmoll -1 MgCl 2 )...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com