Traceless cloning and reorganizing method by means of activity of exonuclease
An exonuclease and activity technology, applied in the field of traceless cloning and recombination, can solve the problems of affecting the experimental results, traces of restriction endonuclease sequences, changing the structure and activity of the target protein, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
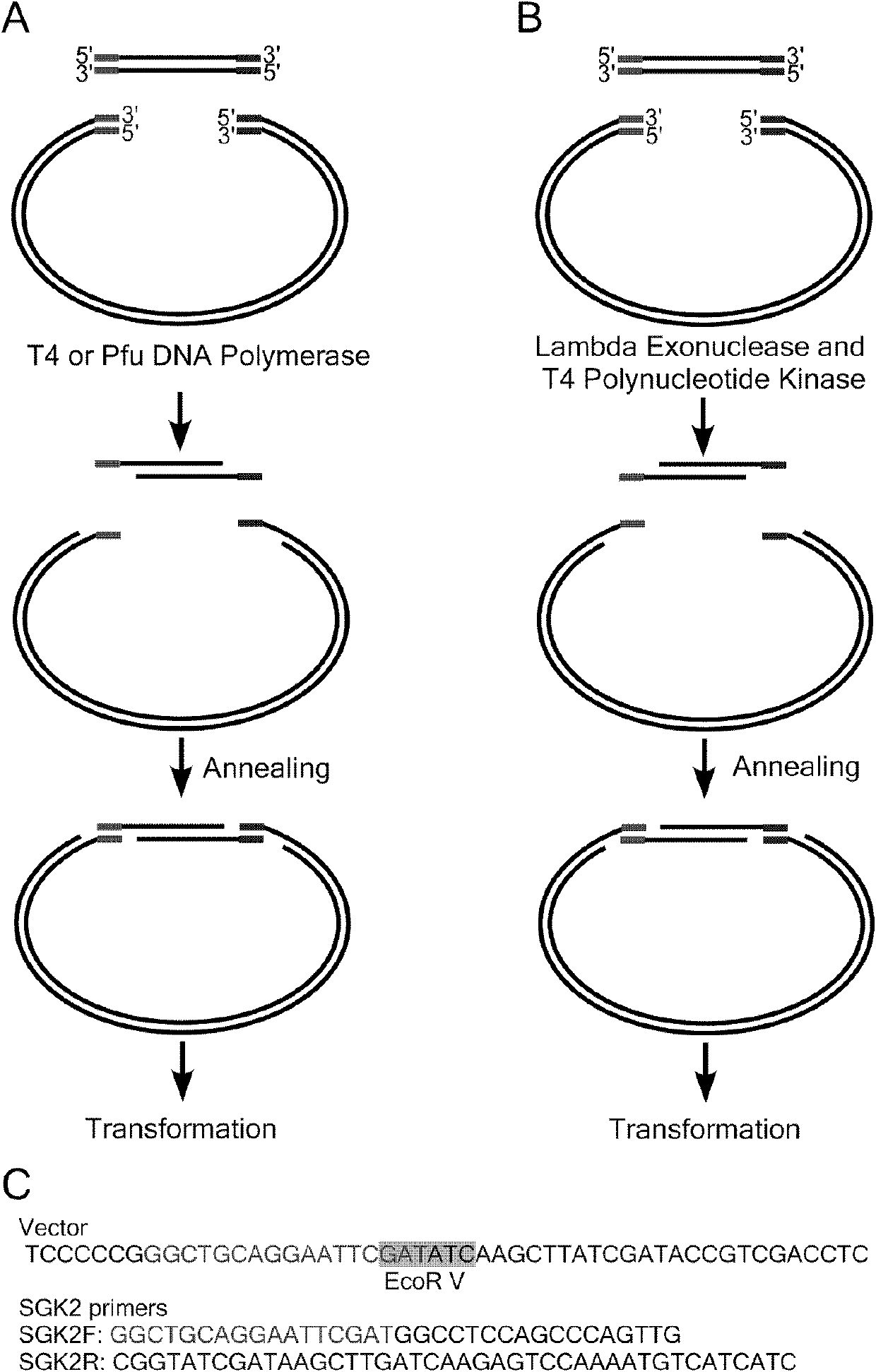
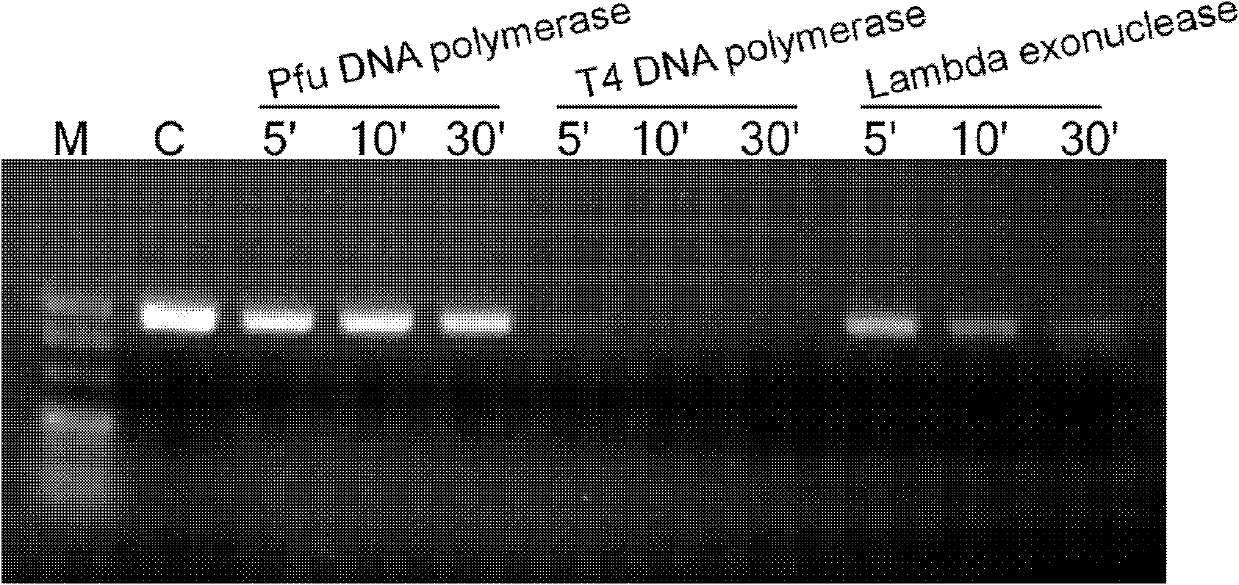
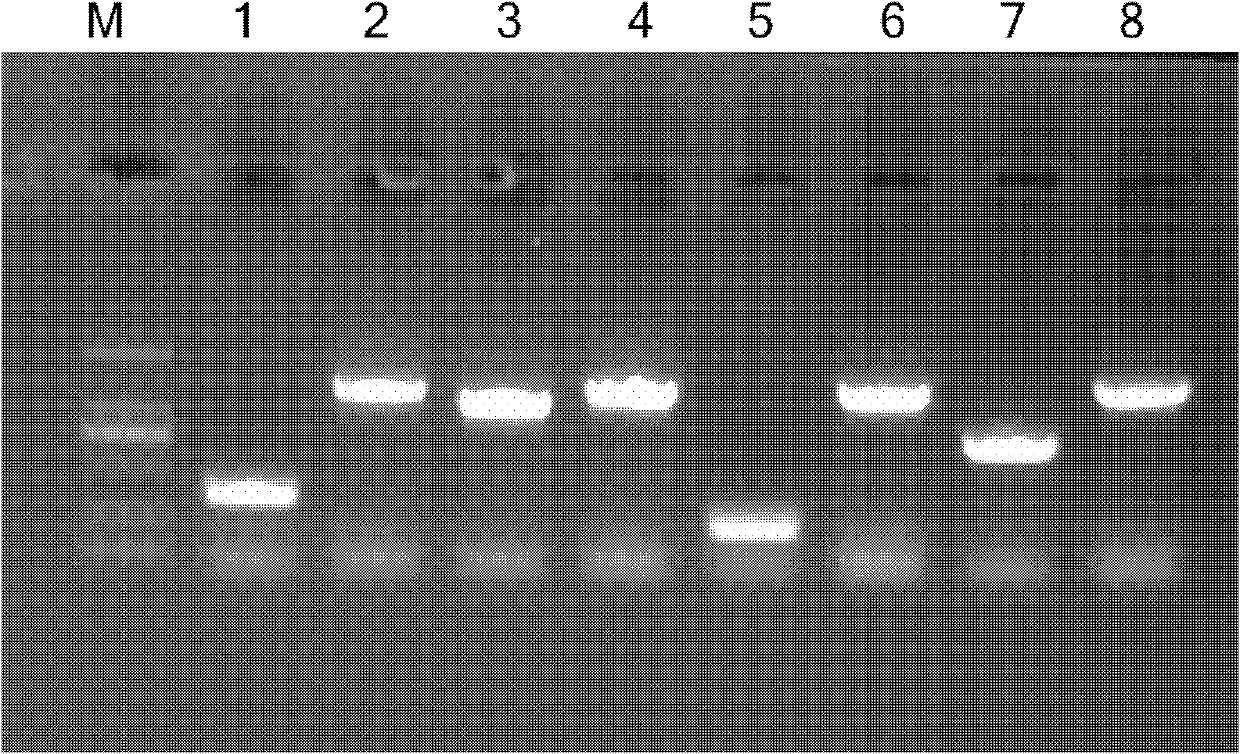
[0029] Example 1: Utilize exonuclease activity to clone the coding region (open reading frame, ORF) of SGK (serum / glucocorticoid regulated kinase 2) gene 7.1, detect the exonuclease activity of Pfu, T4DNA polymerase and lambda Exonuclease
[0030] In a 10 μl reaction system, 100 ng of DNA fragments were reacted with 1 U of Pfu DNA polymerase, T4 DNA polymerase, and λExonuclease (Table 1) for 5 min, 10 min, and 30 min, respectively, and then 2 μl of 50 mM EDTA was added to terminate the reaction and electrophoresis was performed.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com