Triple-fluorescence PCR (Polymerase Chain Reaction) detection method and kit for Nosema bombycis (Nb), Bombyx mori Nuclear Polyhedrosis Virus (BmNPV) and Bombyx mori Densovirus (BmDNV), as well as primers and probes
A technology for karyotype polyhedrosis and microsporidia, which is applied in the field of biological detection technology, can solve the problems of unstable serological detection, low specificity and sensitivity, and inability to simultaneously differentiate and diagnose.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Design of primers and probes
[0044] Based on the highly species-specific microsporidia small subunit ribosomal RNA gene sequence of the silkworm ( Nosema bombycis small subunit ribosomal RNA, FJ854545 (gene sequence number)), silkworm nuclear polyhedrosis virus DNA polymerase gene sequence ( Bombyx mori nucleopolyhedrovirus gene for DNA polymerase, D16231) and the hypothetical structural protein gene sequence of the silkworm densovirus Zhenjiang strain ( Bombyx mori densovirus isolate Zhenjiang putative structural protein gene, AY236978), using bioinformatics methods based on primer design principles, and using Primer Express design software, designed several sets of specific primers and fluorescent probes for amplifying the above gene fragments. The designed primers and probes were verified by BLAST (http: / / blast.ncbi.nlm.nih.gov / ) to ensure the high specificity of the primers. Furthermore, DNAStar software was used to verify whether primer-dimers we...
Embodiment 2
[0048] Example 2: Sample DNA Extraction
[0049] Select 0.2 g of the silkworm material infected with the above three pathogens, break it by glass bead crushing method, 6000 r / min, 20 s×6 times, use the DNA extraction kit (Promega FF3750) to carry out DNA extraction according to the instructions. The extracted DNA was dissolved in 20 μl of water.
Embodiment 3
[0050] Example 3: Triple fluorescent PCR amplification reaction and fluorescence compensation reaction
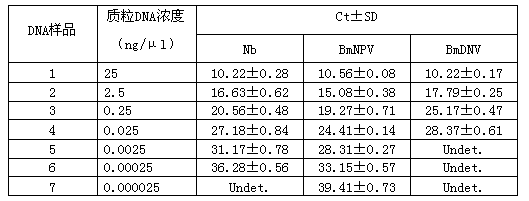
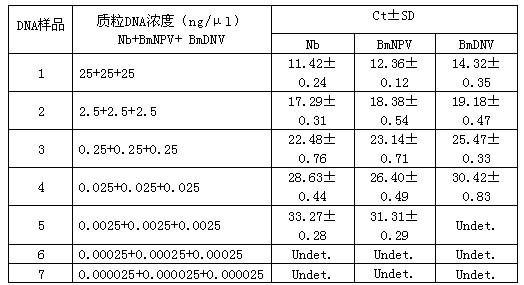
[0051] Simultaneous detection of three samples by triple fluorescent PCR amplification method. The sequences of primers and probes are shown in Table 1. The triple fluorescent PCR reaction system was as follows: 10 μl Premix Ex Taq (Takara), 0.13 μl of each primer (10 pmol / μl), 0.27 μl of each probe (10 pmol / μl), Take 4 μl of the DNA extracted above (the DNA content of Nb, BmNPV and BmDNV is 2.5 ng / μl, respectively), and make up the insufficient volume to 20 μl with water. The fluorescence quantitative PCR instrument lightcycle 480 (Roche) was used for the reaction, and the reaction program was (1) 95°C, 10 sec; (2) 95°C, 5 sec; 60°C, 23 sec; 40 cycles. Call the corresponding fluorescence compensation program, perform fluorescence compensation, and judge the results. It is generally believed that when the Ct value is less than or equal to 35, the result is positive, whe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com