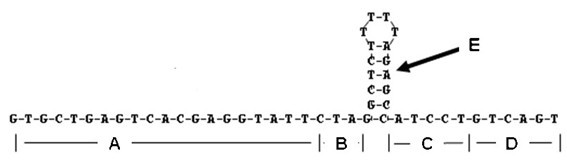
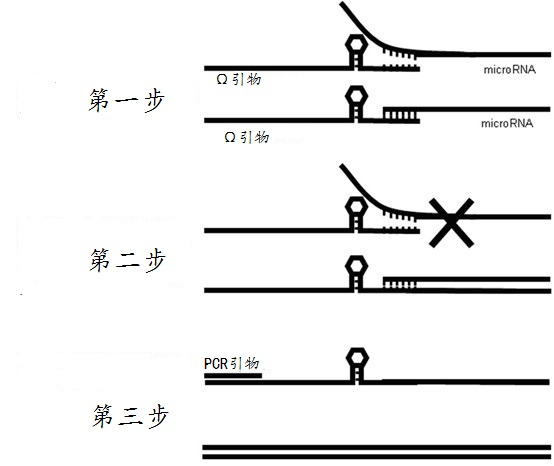
Omega structure oligonucleotide primer for detecting short chain ribonucleic acid (RNA) and application thereof
An oligonucleotide, omega technology, applied in DNA/RNA fragments, recombinant DNA technology, biochemical equipment and methods, etc., can solve problems such as intractability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
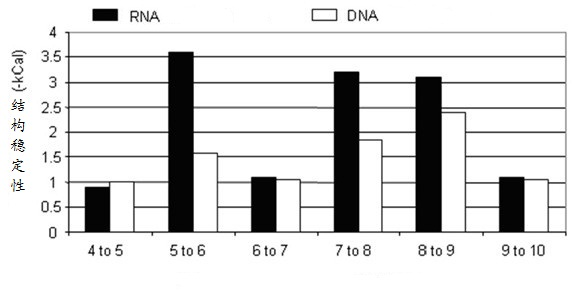
[0072] Embodiment 1: Comparison experiment of RNA oligomeric chain and DNA oligomeric chain stability
[0073] Such as image 3 As shown, RNA with a 6-paired base stem and DNA with a 9-paired base stem are the most economical structures. The backbone chains of RNA and DNA are different, and the energy contribution to maintain the secondary structure is also different. As the paired nucleotides in the stem region increase, the Gibbs free energy decreases. However, the contribution of each added base pair to structural stability varies according to its position. For example, adding one base pair to a 4-base convex-stem RNA reduces the free energy by -0.9 kcal / mole (kcal / mole mole), a 5-base convex-stem RNA adds one base pair, and the free energy drops by -3.6 kcal / mole. The 6-base-pair convex-stem RNA is the most economical structure, which yields the greatest free energy gain with the least number of nucleotides. The 6-base pair convex stem structure is the most common RN...
Embodiment 2
[0074] Embodiment 2: the functional experiment of the probe spacer of omega primer of the present invention
[0075] A sequence in the chain of TPL6 designed and synthesized is the same as its 3' end sequence as a template.
[0076]
[0077] TPL6 sequence (complementary strand): SEQ ID NO:2
[0078]
[0079] RT reactions were simulated with omega primers with 8 base pair stem lengths, and nucleotides in italics were used as probe spacers.
[0080]
[0081] Dissolve RT primers with 1x TE buffer at a concentration of 50 μM. Add 1 μl of primers to 500 μl H2O dilution, and use 1 μl of diluted primers per RT reaction. Perform 10-fold serial dilutions of TPL6 with sterilized water, dilution 10 7 – 10 12 as a template. The 5 μl RT reaction system consisted of 1x PCR buffer, 0.1xTE, 50 nM omega primer, 0.05 u / μl Taq DNA polymerase and different dilutions of TPL6 template. The conditions of the RT reaction were: 37°C for 10 minutes, 20°C for 1 minute and 37°C for 10 sec...
Embodiment 3
[0086] Example 3 Contrastive experiment of the influence of probe region length on reverse transcription efficiency and in-chain priming
[0087] The probe types used are:
[0088] A: Stem-loop primers + probes indicated for each plate below
[0089] B: Linear primers + probes as indicated for each plate below
[0090] C: Omega primers + probes indicated for each plate below
[0091] The TLP5 template sequences used for plates 1-5 were (complementary strand):
[0092]
[0093] The primer probe sequences are as follows:
[0094] Board 1. AGT
[0095] Board 2. AGTC
[0096] Board 3. AGTCA
[0097] Plate 4. AGTCAG
[0098] Plate 5. AGTCAGT
[0099] Template TLP1 used for plate 6 - no intrastrand site, the sequence is (complementary strand):
[0100]
[0101] The probe sequences are:
[0102] Plate 6. AGTCAGT
[0103] Primers were dissolved in 1xTE at a concentration of 50 μM, denatured at 95°C 10”, kept at 65°C for 5’, and then cooled to room temperature. Add 1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com