Long genome segment deletion system and use thereof
A gene and fragment technology, applied in the direction of recombinant DNA technology, hydrolytic enzymes, etc., can solve the problems of poor robustness and economic traits, difficulty breaking through the limitation of variety resources, and single genetic background, and achieve accurate genetic transformation, high genetic transformation efficiency, The effect of enriching genetic resources
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
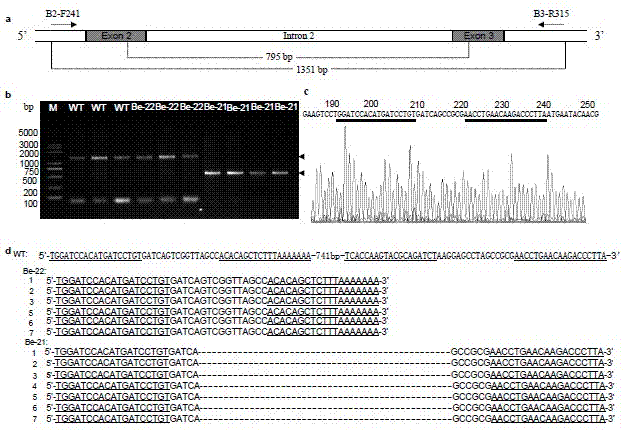
[0025] Example 1. Silkworm oil silkworm gene BmBlos2 Sequence analysis
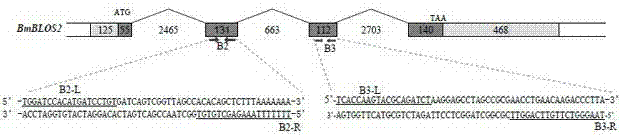
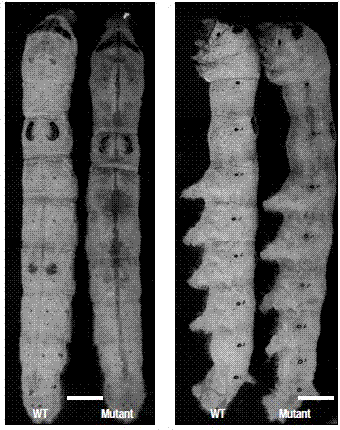
[0026] Download silkworm oil silkworm gene from silkDB database BmBlos2 The sequence (numbered BGIBMGA002101). Sequence analysis showed that the gene includes four exons of 180 bp, 131 bp, 112 bp and 608 bp and three introns of 2465 bp, 663 bp and 2703 bp. The first exon includes a 125 bp untranslated region and 55bp coding region, its structure is as figure 1 Shown. According to the general principle of gene knockout, the second and third exons of the gene are ideal target sites. In this embodiment, the second and third exons are the target sites.
Embodiment 2
[0027] Example 2. The third exon nuclease TALEN -B3 the design of
[0028] According to Bombyx mori gene BmBlos2 The sequence characteristics of the third exon, we chose the sequence on the third exon as the target sequence of the transcription activator-like effector nuclease TALEN-B3, the silkworm oil silkworm gene BmBlos2 The nucleotide sequence of the third exon is shown in SEQ ID NO. 1. According to the TALEN principle, the nucleotide sequence of the structure T(Nn)A is the target sequence recognized by the transcription activator-like effector nuclease TALEN, where N It is any base of A, G, T and C, and n is any number between 24 and 65. According to the above principles, the target site recognized by TALEN is designed to be 5’- tcaccaagtacgcagatcta aggagcctagccgc g aacctgaacaagaccctta -3'(SEQ ID NO.2), the underlined part is the recognition module of the transcriptional activator-like effector nuclease TALEN-B3, the bold part represents the spacer sequence, and the u...
Embodiment 3
[0030] Example 3. Design of the second exon nuclease TALEN-B2
[0031] According to Bombyx mori gene BmBlos2 The sequence characteristics of the second exon were selected as the target sequence of TALEN-B2, the silkworm oil silkworm gene BmBlos2 The nucleotide sequence of the second exon is shown in SEQ ID NO.5. According to the TALEN principle, the nucleotide sequence of the structure T(Nn)A is the target sequence of TALEN, where N is A, G, T and C In any base, n is any number between 24 and 65. According to the above principle, the target sequence designed for TALEN is 5’- tggatccacatgatcctgt gatcagtcggttagcc acacagctctttaaaaaaa -3'(SEQ ID NO.6), the underlined part is the TALE recognition module, the bold part represents the spacer sequence, and the underlined nucleotide sequence at the 5'end is the 5'end recognition module, corresponding to SEQ ID NO.2 Nucleotides 1-19 of the 3'end of the nucleotide sequence is the 3'end recognition module, corresponding to the 36-54 sh...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com