Primer and kit for detecting coxsackievirus A6 type RT-LAMP (Reverse Transcription Loop-mediated Isothermal Amplification) nucleic acid
A technology of RT-LAMP and Coxsackie virus, which is applied in the application field of biological detection technology, can solve the problems of simple, fast and accurate diagnosis, unfavorable on-site sample detection, unfavorable grass-roots promotion, etc., to meet the requirements of rapid on-site detection and low cost Low, fast detection effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1: Design and synthesis of LAMP primers
[0023] According to the CA6VP1 gene sequence published by GenBank, its relatively conserved region was analyzed using the biological software CLAST, and the LAMP online design software PrimerExplorer V4 ( http: / / primerexplorer.jp / e / ) designed six primers for relatively conserved sequence regions, including two inner primers FIP (F2+F1C) and BIP (B2+B1C), two outer primers F3 and B3, and two loop primers LF and LB, respectively Eight binding regions in the target sequence were matched, and the primers were evaluated using Oligo 6 software and BLAST.
[0024] The primer sequences are:
[0025] F3 (Forward Outward Citation): 5'-ACTCGCTGTGTGATGAATCG-3'; that is, SEQ ID NO:1
[0026] B3 (reverse external reference): 5'-GCGTTGTGCTATCATTGAGG-3'; ie SEQ ID NO:2
[0027] FIP(F1C+F2, forward introductory):
[0028] 5'-CCTTCACCTCCACAACTCCTACTGAGGCGAGTGTGGAACA-3'; namely SEQ ID NO:3
[0029] BIP (B1C+B2, reverse internal citati...
Embodiment 2
[0037] Embodiment 2: the extraction of RNA
[0038] The RNA extraction of virus samples uses Roche High Pure Viral RNA Kit, and the steps are carried out according to its operating instructions. Specific steps: 1. Add 400 μl Binding Buffer to 200 μl sample treatment solution, mix well, transfer to a filter column, and centrifuge at 8000 g / min 15s; 2. Discard the filtrate, replace the collection tube, add 500μl Inhibitor Removal Buffer, centrifuge at 8000g / min for 1min; 3. Discard the filtrate, replace the collection tube, add 450μl Wash Buffer, centrifuge at 8000g / min for 1min; 4. Repeat step 3 once; 5. Discard the filtrate, replace the collection tube, and centrifuge at the maximum speed of the centrifuge for 10s; 6. Transfer the filter column to a 1.5ml EP tube, add 50μl Elution Buffer, and centrifuge at 8000g / min for 1min. The eluate is the purified nucleic acid. Virus cell culture and herpes fluid are directly extracted according to the above method; fecal samples, anal sw...
Embodiment 3
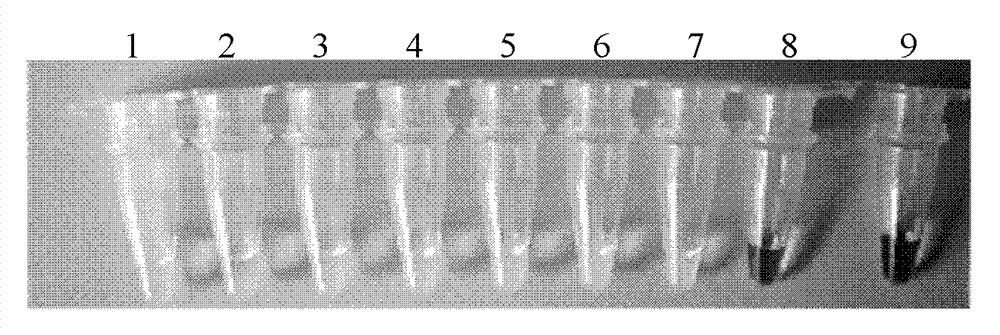
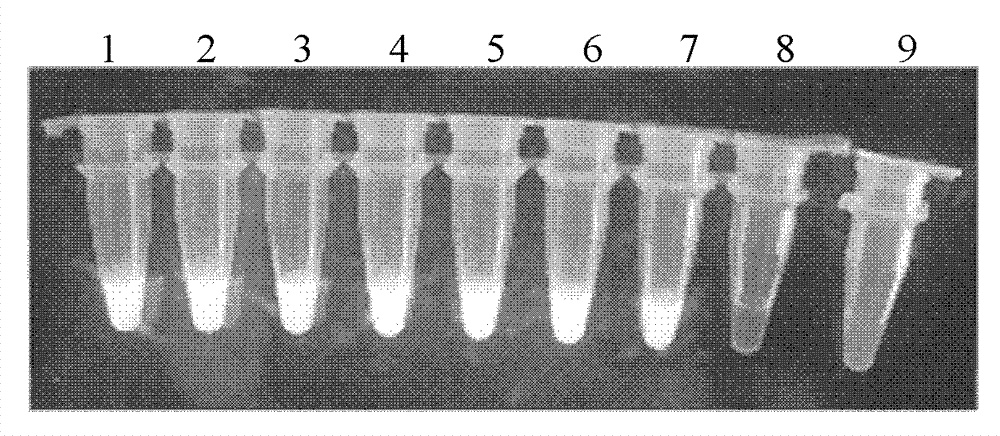
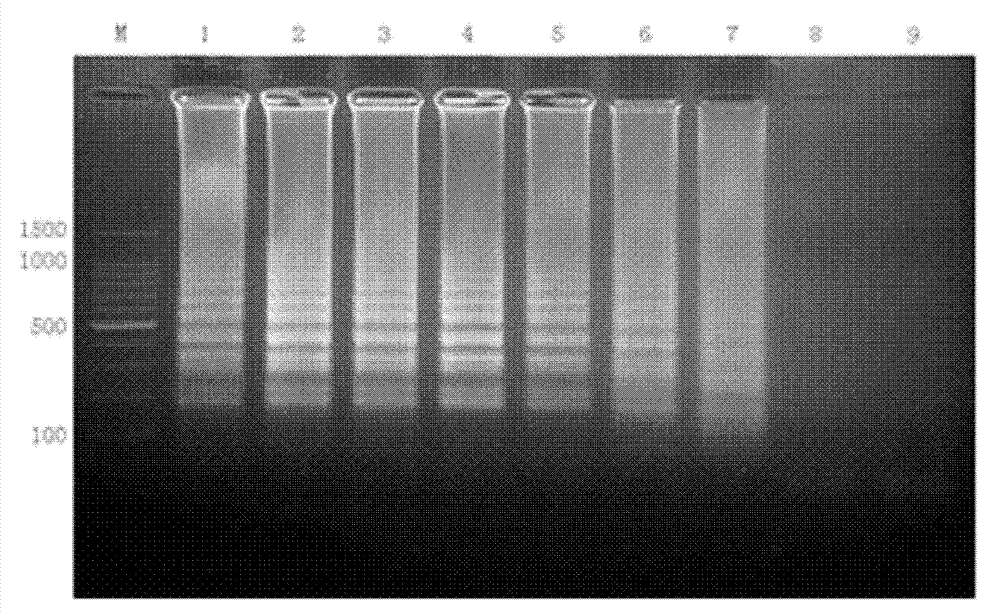
[0039] Example 3: Establishment of CA6RT-LAMP amplification method
[0040] 1. CA6RT-LAMP reaction system
[0041] Use the sample RNA to be tested as a template for RT-LAMP reaction.
[0042] Table 1 CA6RT-LAMP reaction system
[0043] components
volume
RT-LAMP reaction solution
23μl
RNA
2μl
total capacity
25μl
[0044] Wherein, the preparation of RT-LAMP reaction solution is shown in Table 2.
[0045] Formulation of table 2CA6RT-LAMP reaction solution
[0046] components
Final concentration
working concentration
volume
10×buffr
1×
2.5μl
MgCl 2
5mM
25mM each
5μl
dNTPs
1.4mM each
10mM each
3.5μl
FIP
1.6μM
40μM
1μl
BIP
1.6μM
40μM
1μl
F3
0.2μM
10μM
0.5μl
B3
0.2μM
10μM
0.5μl
LF
0.8μM
40μM
0.5μl
LB
0.8μM
40μM
0.5μl
AMV
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com