Method for finding E2-E3 specifically mediating target protein ubiquitination reaction based on known E1
A target protein, specific technology, applied in the field of biochemistry and molecular biology, can solve the problems of terror and unclear ubiquitin modification mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
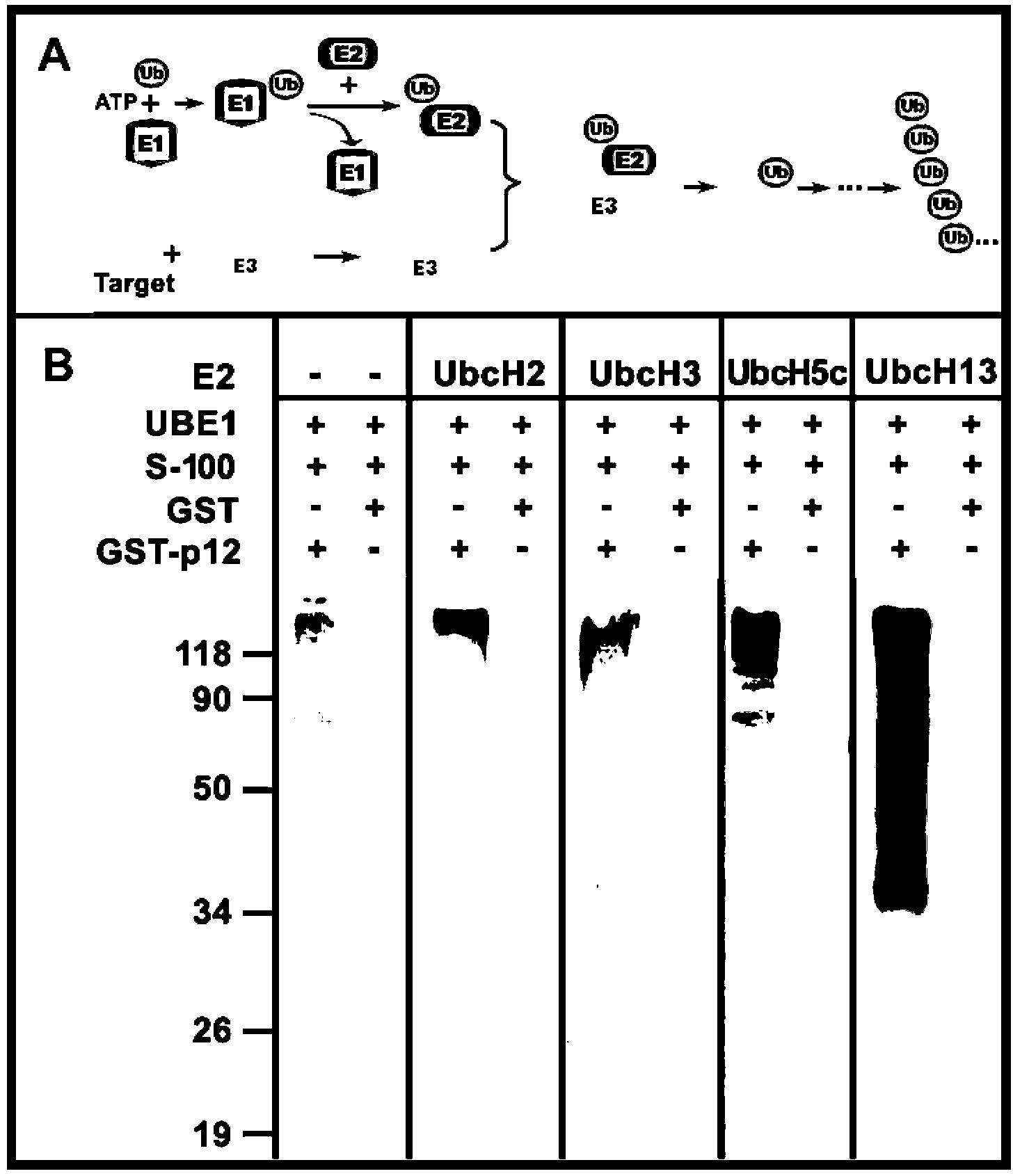
[0035] Example 1: Establishment of in vitro reaction system for ubiquitination modification and determination of E2
[0036] A 15 μL reaction system was used for in vitro ubiquitination reaction, containing: 10 μg ubiquitin, 30 nM UBE1, 20 ng of Ubiquitin aldehyde, 1x ENS, 300 ng GST-p12 as substrate, HeLa S-100 fraction as E3 source, and 500 nM UbcH5c, UbcH2, UbcH3(Cdc34) and UbcH13 / Uev1a complex as E2 respectively; reaction buffer is 5 mM MgCl 2 , 40 mM Tris-HCl, pH 7.5, 2 mM DDT, and GST was used as a control experiment. at 30 0 React under C condition for 60 minutes. At the end of the reaction, add 800 μL of Pull-down buffer (1xPBS containing 0.5% NP-40) to terminate the reaction, then add 10 μL of Glutathione Sepharose-4B beads, and shake at room temperature for 60 minutes; Wash the beads 6 times with Pull-down buffer; add 1 x SDS-PAGE loading buffer, 95 0 C for 5 minutes, and the samples were electrophoresed on a 12% SDS-PAGE gel and then transferred to the membrane. ...
Embodiment 2
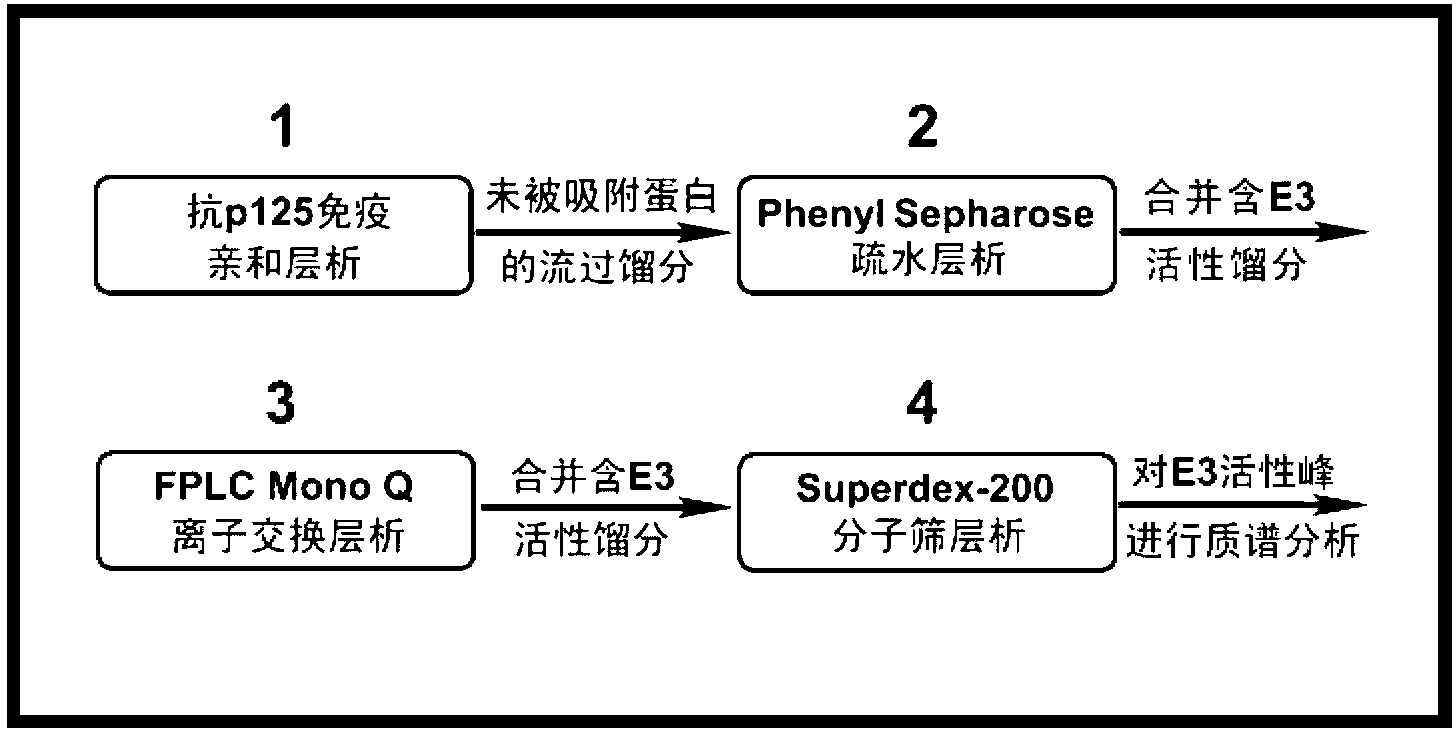
[0037] Example 2: Determination of the E3 activity of each fraction after the first-stage immunoaffinity chromatography
[0038] about 2×10 8 Hela cells cultured in vitro, lysed by ultrasound, 4 0 After high-speed centrifugation at C, take the supernatant as the cell lysis extract (the cell lysis buffer used is: 20 mM Tris-Cl, pH7.8, 0.5 mM EGTA, 1 mM EDTA, 1 mM MgCl, 50 mM NaCl , 10% Glycerol, Protease Inhibitor). After such figure 2 The schematic diagram of the technical route shown, through the combination of four chromatographic columns to identify unknown E3. Detect the E3 activity of the eluted fractions in each step of chromatography, and then combine the E3 activity peaks and pass through the next chromatography column. After the last step of chromatography, conduct mass spectrometry identification on the fraction peaks containing E3 activity to determine the specific recognition target E3 of protein p12.
[0039] Taking UBE1 as E1 and the UbcH13 / Uev1a complex de...
Embodiment 3
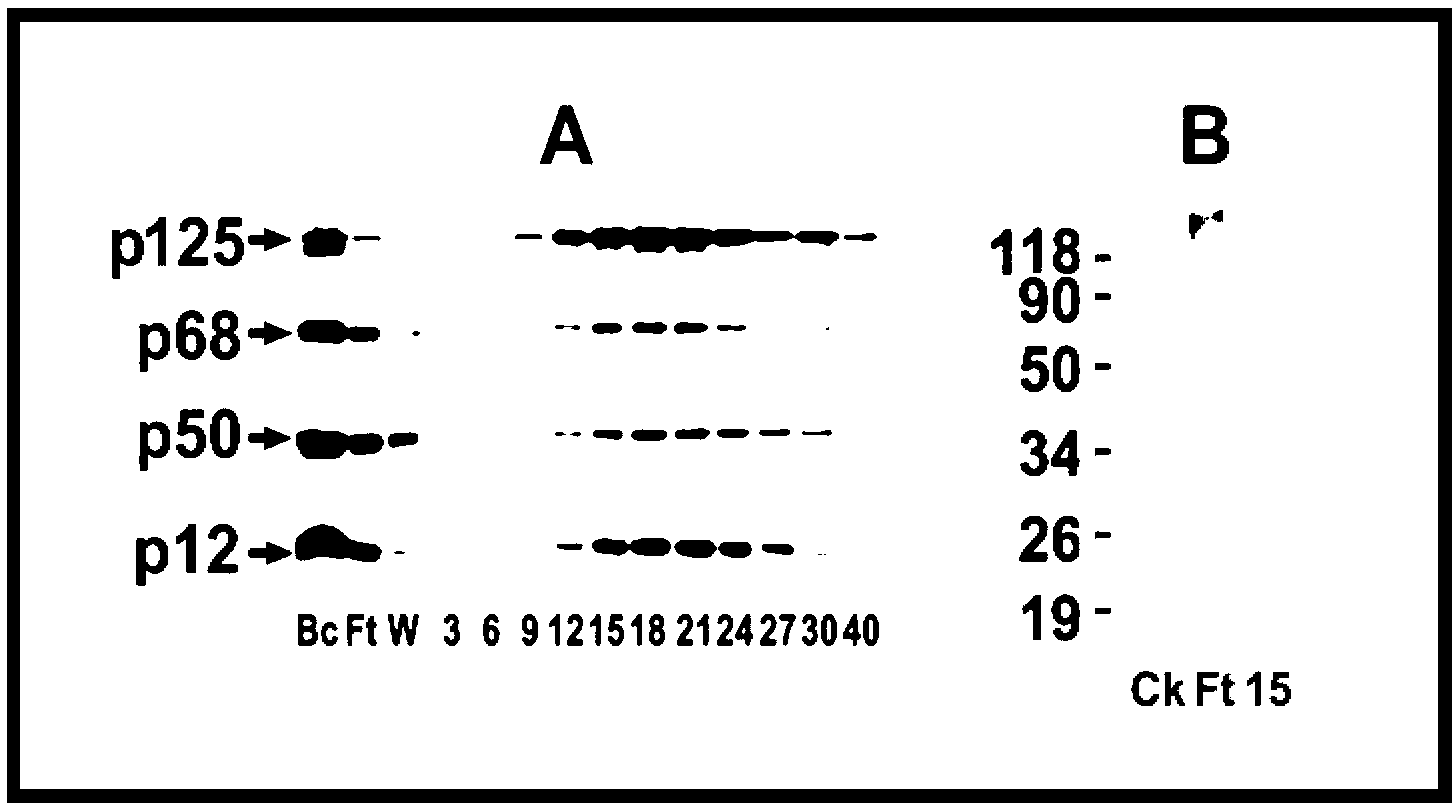
[0040] Example 3: Determination of the E3 activity of each fraction after separation by the second-stage Phenyl Sepharose hydrophobic chromatography
[0041]The Ft in Example 2 was dialyzed in the dialysate TGEED (TGEE+1 mM DDT) / 500 mM ammonium sulfate, and the dialyzed Ft was separated by Phenyl Sepharose hydrophobic chromatography, and TGEED containing 500 mM to 0 mM ammonium sulfate for gradient elution, and then detect the E3 activity in each eluted fraction. Such as Figure 4 As shown, the E3 activity using GST-p12 as substrate mainly exists in fraction No. 50. Fractions 40-60 were pooled and dialyzed against TGEED dialysate to remove ammonium sulfate.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com