Method for constructing multi-copy kluyveromyces lactis expression vector efficiently in in-vitro manner
A Kluyveromyces cerevisiae and expression vector technology is applied in the field of efficient construction of multi-copy Kluyveromyces lactis expression vector in vitro, which can solve the problems such as the inability to meet the yield requirement, the expression amount and the stability to be improved, and achieve the improvement of protein expression. Expression level, the effect of increasing copy number
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
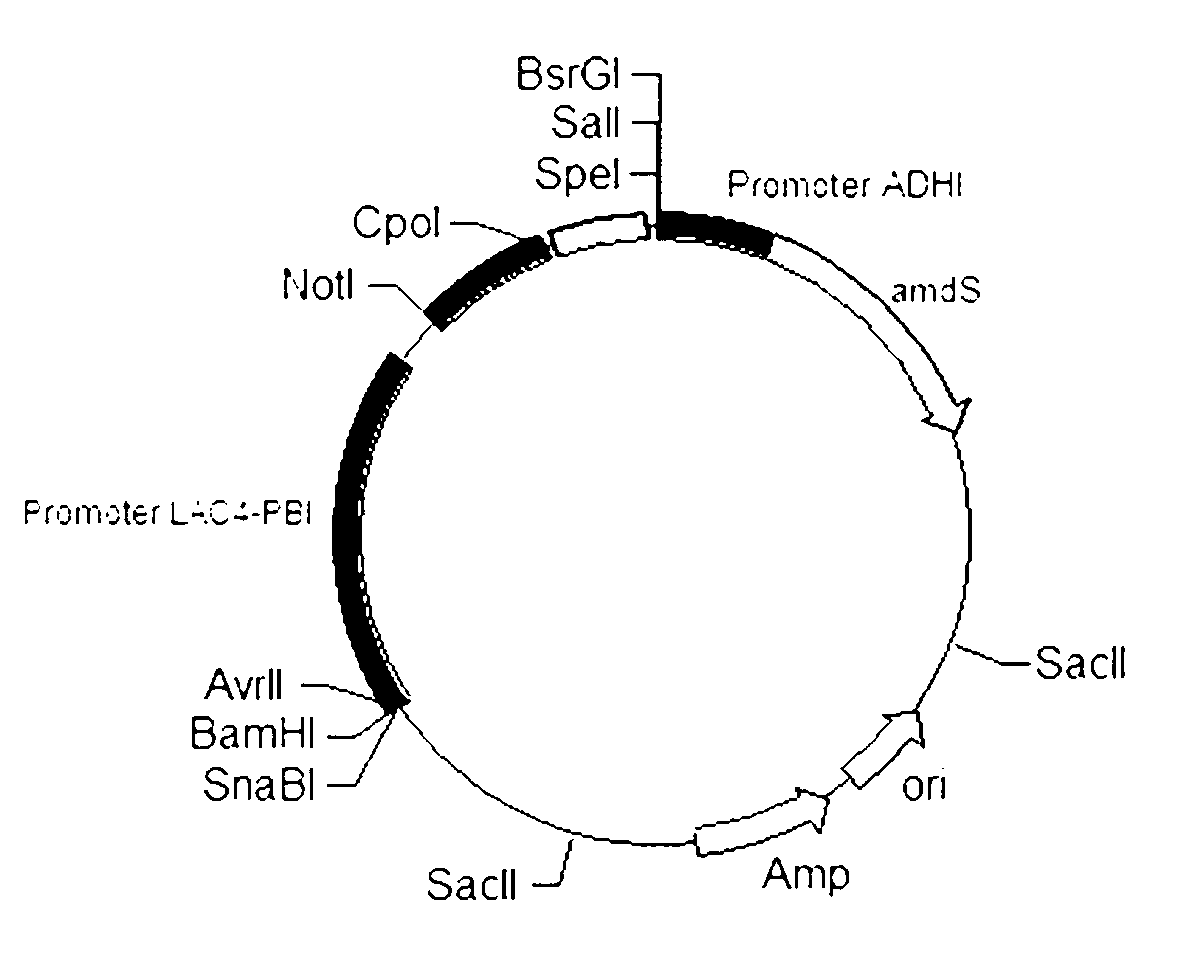
[0025] Example 1 Build in the above way Expression vector PPML 2 .
Embodiment 2
[0026] Example 2 Construction of the recombinant vector GG799-Bp-Bman containing the mannanase gene.
[0027] ,choose Bacillus subtilis ( Bacillus subtilis ) derived mannanase gene is the target gene;
[0028] 2、 Synthetic primers. The primers in Table 2 were designed with GeneTool software and synthesized for later use.
[0029] Primer name sequence Bman-f 5'atagcggccgcAAACCAATTGAAGCGCATACTGTGT 3 Bman-r 5'atacggtccgTTACTCAACGATTGGCGTTAAAAGAATC 3
[0030] sheet
[0031] 3. Construction of expression vector. Bacillus subtilis ( Bacillus subtilis) derived mannanase gene with primers Bman-f and Bman-r for PCR (98°C, 2min, 1 cycle; 98°C, 7s, 55°C, 30s, 72°C, 1min, 25cycle; 72°C, 8min, 1 cycle; stored at 4°C), and the product was recovered by agarose gel electrophoresis and sequentially treated with restriction endonuclease not I and Cpo Ⅰ Enzyme digestion, after recovery, connect to not I and Cpo Ⅰ Expression vector BpKLAC after ...
Embodiment 3
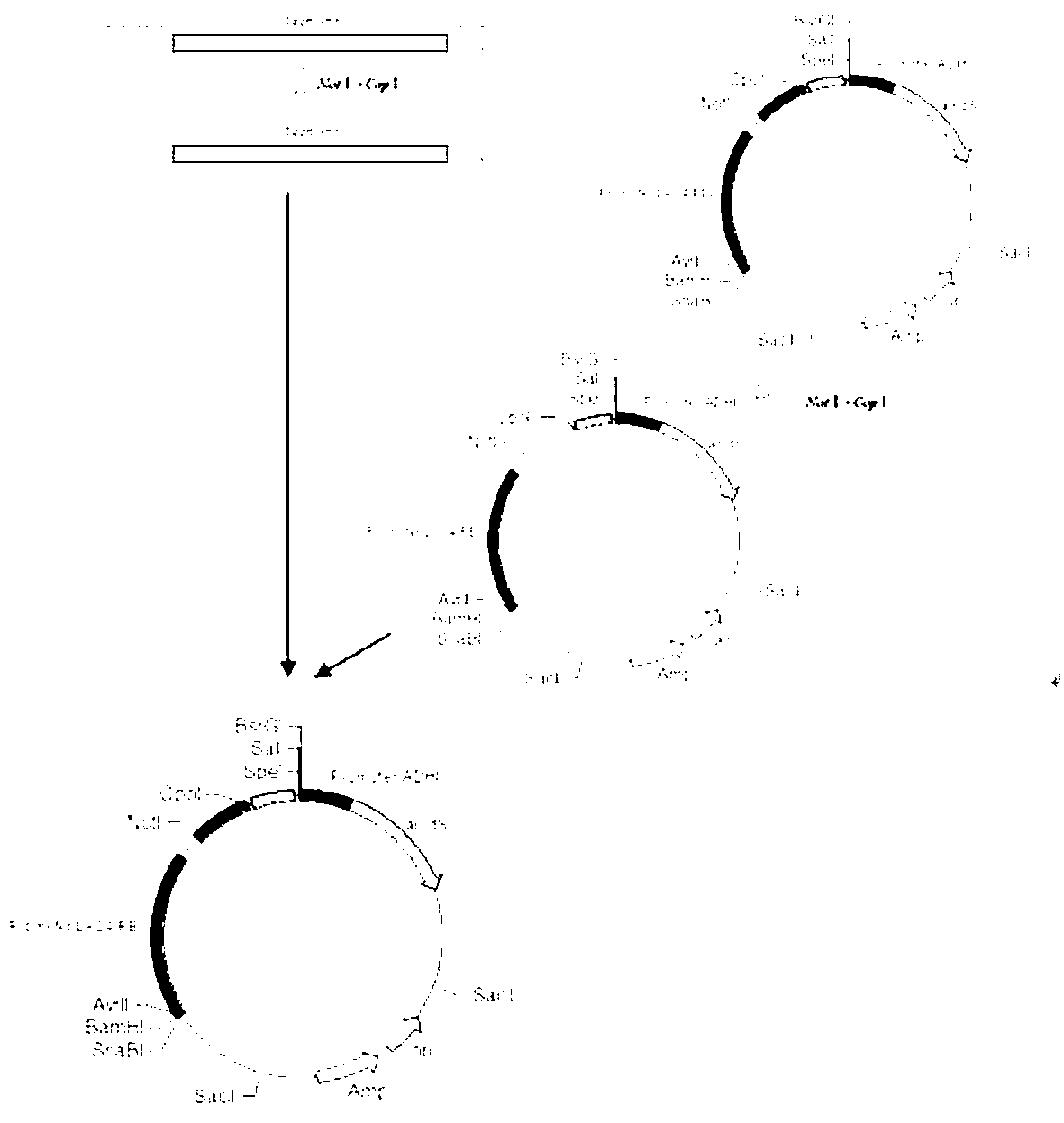
[0035] 1. Using restriction endonuclease Bam H I and Speech Ⅰ Double digestion instance 2 vectorBpKLAC 2 -manB, resulting in a biobrick body containing cohesive ends, named Fragment 1. restriction endonuclease BIn I and Sal Ⅰ Double digestion vector BpKLAC 2 -manB, to obtain the Biobrick body containing cohesive ends, named Fragment 2, with restriction endonucleases Bam H I and Sal Ⅰ Double digestion vector BpKLAC 2 -manB, resulting in biobrick vector fragment 3 containing cohesive ends.
[0036] Mix Fragment 1, Fragment 2 and Fragment 3 at a ratio of 3:3:1 (1.5 μl for Fragment 1 and 2, and 0.5 μl for Fragment 3), use TAKARA’s ligation kit Solution I ligase for 2 hours, and routinely transform into Escherichia coli competent cells DH10β were coated on LB plates containing ampicillin sodium and cultured at 37°C for 12 hours. Randomly pick colonies, extract plasmids, and use restriction endonucleases Bam H I and Sal Ⅰ Double enzyme digestion to verify whethe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com