Library preparation method of trace nucleic acid sample and application thereof
一种核酸文库、核酸样本的技术,应用在微量核酸样本的文库制备领域,能够解决阻碍单细胞和微量核酸应用等问题
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
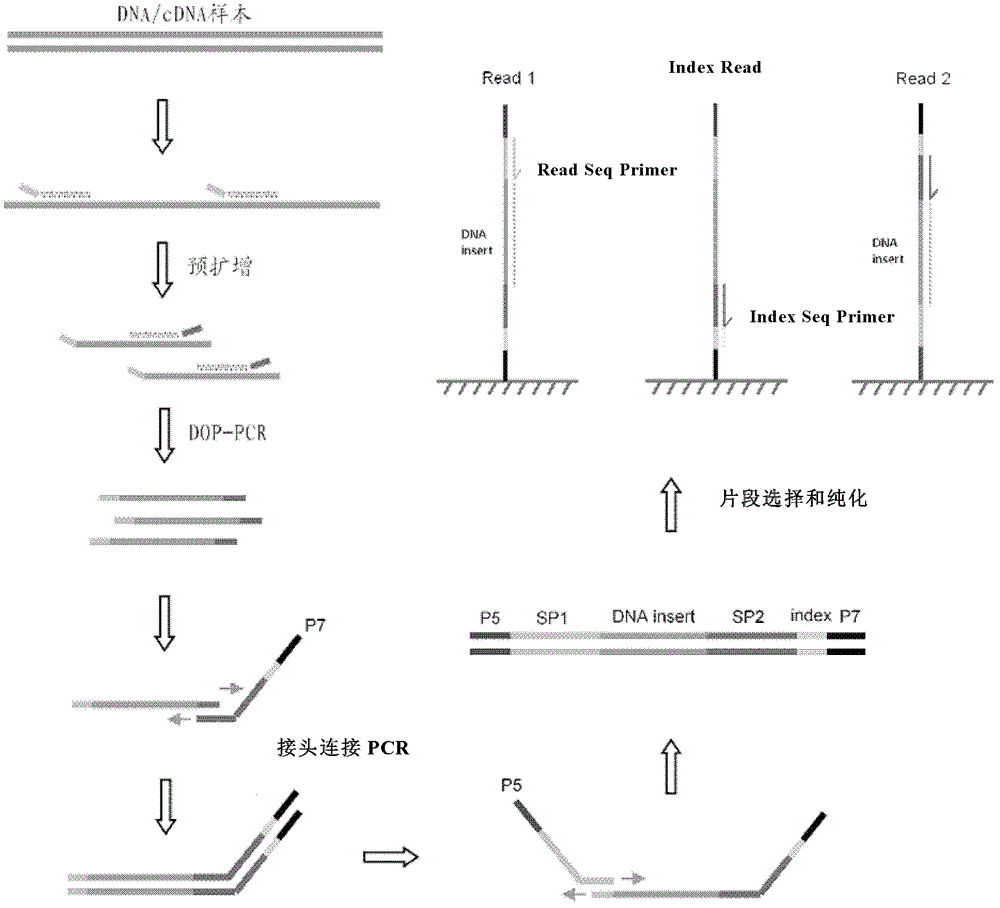
[0104] Preparation of single-cell genomic DNA library by DOP-PCR method
[0105] In this embodiment, a single lymphocyte in the blood is taken as an example, and a single-cell genomic DNA library is prepared by the DOP-PCR method.
[0106] 1. Sample source:
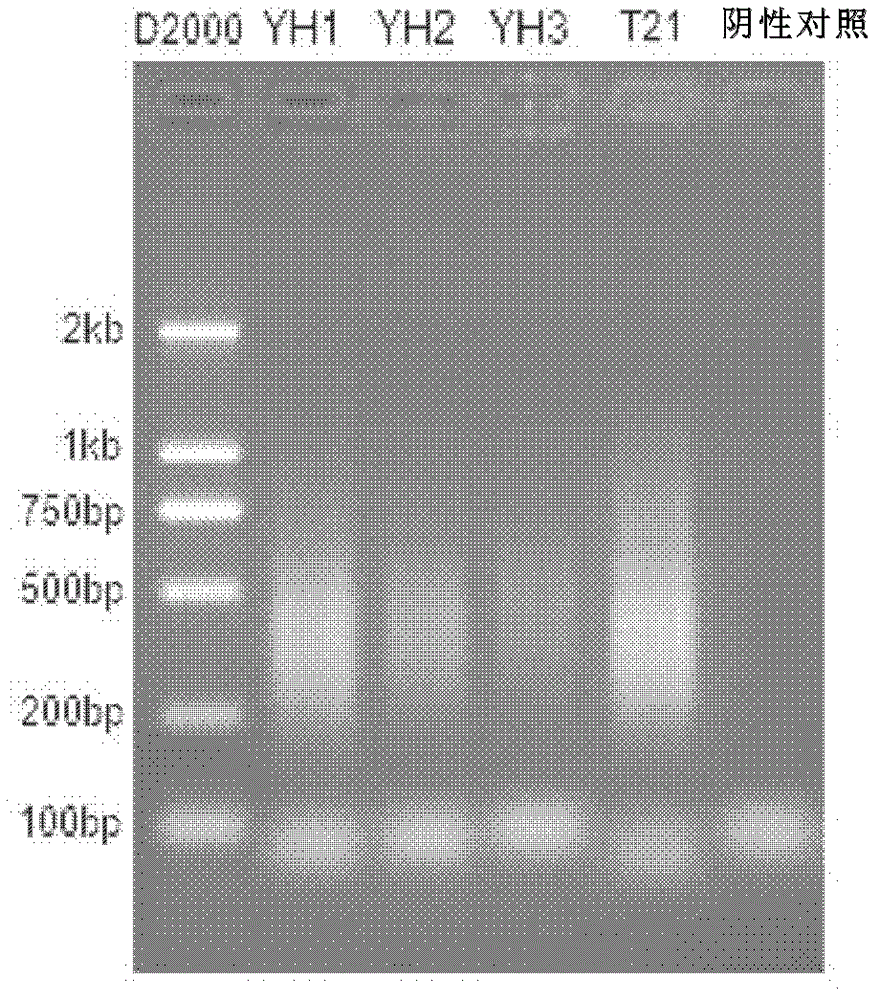
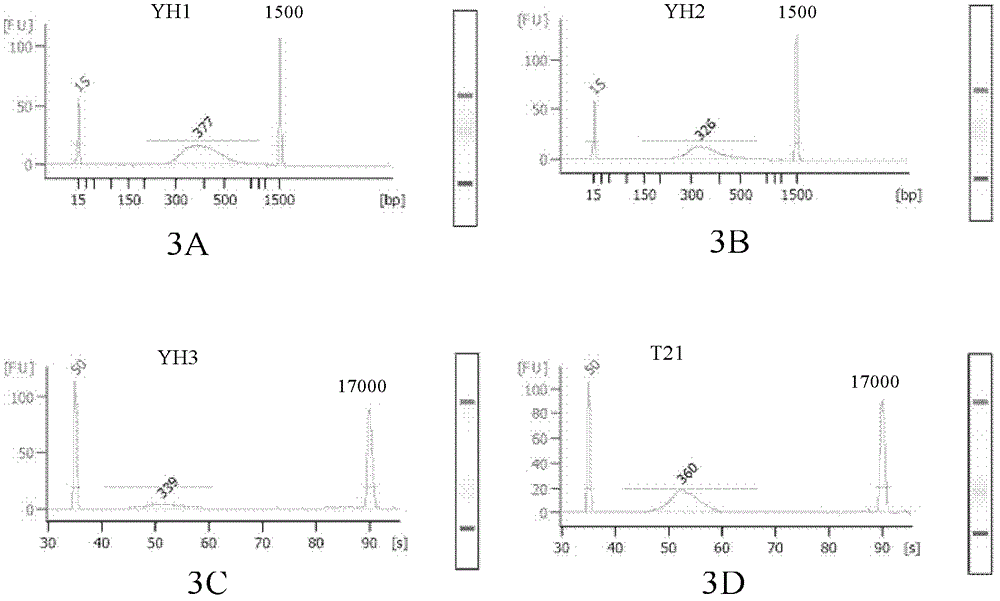
[0107] The blood single cell samples came from the peripheral blood of a normal person (YH, Yanhuang Project sample) and a patient with Down syndrome (T21).
[0108] 2. Single cell separation:
[0109] A total of 3 YH and 1 T21 peripheral blood single lymphocytes were separated by mouth pipette method and placed in a PCR tube containing 2 μl alkaline cell lysate (200 mM KOH, 50 mM DTT) and stored at -80° C. for more than 30 minutes.
[0110] 3. The first PCR (low stringency amplification, DOP pre-amplification):
[0111] DOP pre-amplification The PCR tube containing single cells was treated at 65°C for 15 minutes, and then DOP reaction solution was added to perform DOP pre-amplification reaction. In this example, pfx DNA polymerase ...
Embodiment 2
[0168] Repeat Example 1, the difference lies in the DOP pre-amplification reaction conditions, samples and other experimental conditions are the same. The DOP pre-amplification reaction conditions are shown in Table 8.
[0169] Table 8
[0170]
[0171] The results show that the DOP pre-amplification reaction conditions of this example can also achieve the purpose of constructing a library and using it for the next step of detection.
Embodiment 3
[0173] DOP-PCR method for preparing library of trace DNA / cDNA samples
[0174] The sample in this embodiment is a trace DNA / cDNA sample, including trace genomic DNA, immunoprecipitation (IP) product DNA, plasma free DNA (Plasma DNA), and RNA reverse transcription cDNA product.
[0175] The samples of IP product DNA, plasma free DNA (Plasma DNA), and genomic DNA (gDNA) were all diluted 5-fold, and the library was prepared starting with 200pg, 40pg, and 8pg respectively.
[0176] The cDNA sample was obtained by reverse transcription of 1μg of total mouse RNA with hexanucleotide random primers by Superscript II reverse transcriptase, and was diluted 5-fold to the original concentration and 5 -1 , 5 -2 , 5 -3 The concentration starts with library preparation.
[0177] A small amount of DNA / cDNA sample is directly added to the reaction solution to perform the DOP pre-amplification reaction. This embodiment still uses pfx polymerase as an example.
[0178] The reaction system was prepared acc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com