Pig catenin alpha-like 1 (CTNNAL1) gene as genetic marker of pig litter size traits
A genetic marker and catenin technology, applied in genetic engineering, plant gene improvement, recombinant DNA technology, etc., can solve problems such as long time, no discovery, and difficulty in establishing resource families
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Obtaining pig CTNNAL1 gene fragments and establishing a polymorphism detection method
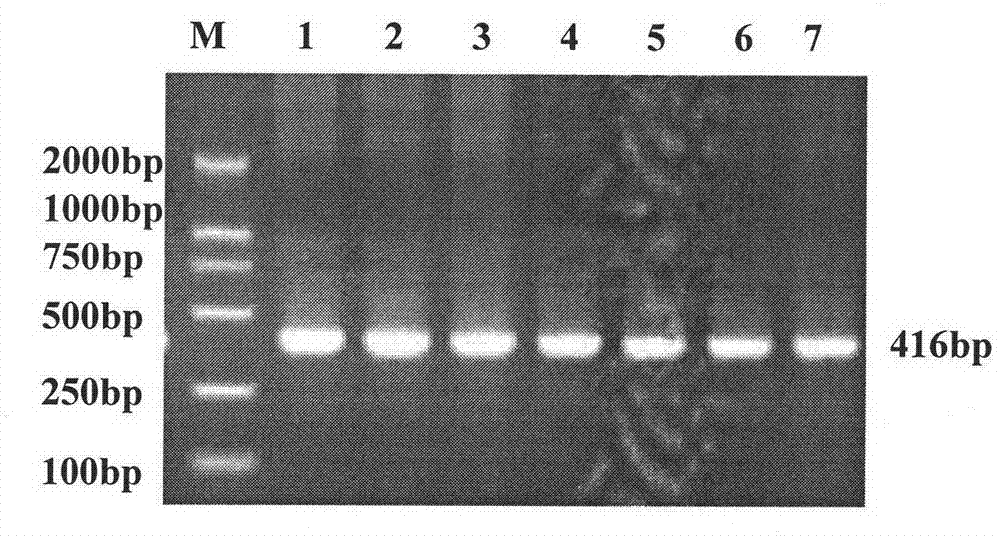
[0031] Log in to the NCBI database to download the corresponding human gene sequence, use the human CTNNAL1 mRNA sequence as the seed sequence, and use the NCBI website EST-others to search for homologous ESTs of pigs. Select the porcine ESTs with more than 80% homology, use electronic cloning, splice the obtained ESTs, and use this as the target sequence, and use Primer2 software to design primers online. The primer sequences are shown in SEQ ID NO: 4 and SEQ ID NO: 5 in the sequence table, as follows: Forward primer CTNNAL1-F: CCATGCTGATCATGTGGTTC, CTNNAL1-R: CAAACCCTCCTCAGCAAAAA. PCR amplification included the fragment of exon 14 of CTNNAL1 gene. The PCR reaction system was 25 μl, in which the template DNA was 50 ng, the concentration of dNTPs was 200 μmol / L, the concentration of each primer was 0.4 μmol / L, 3 U of Taq DNA polymerase (Biostar International, Canada), and...
Embodiment 2
[0033] Example 2: Polymorphic distribution of genetic markers prepared by the present invention in different pig herds
[0034] The extraction of pig genome DNA (sample is shown in Table 1) was carried out with reference to the method introduced in Xiong Yuan's "Introduction to Pig Biochemistry and Molecular Genetic Experiments" China Agricultural Press, 1999.
[0035] In 11 groups of pigs, including 7 foreign blood herds and 3 local Chinese blood herds, 1 synthetic line (Chinese lean pig new line DIV line (co-cultivated by Huazhong Agricultural University and Institute of Animal Husbandry and Veterinary Medicine, Hubei Academy of Agricultural Sciences) , a new breed of lean-meat pig that was popularized and applied on a large scale in China) to detect the PCR-AluI-RFLP polymorphism of pig CTNNAL1 gene, and the detection results are shown in Table 1. The results show that only in large white pigs, Hubei white pigs DIV and Huainan pigs There were 3 genotypes in ; however, no CC...
Embodiment 3
[0039] Example 3: Association analysis of genetic markers prepared by the present invention and litter size traits
[0040] In order to determine whether PCR-AluI-RFLP of pig CTNNAL1 gene is related to pig phenotype differences, 120 Chinese lean pig new strain DIV lines established by the Key Laboratory of Pig Genetics and Breeding of the Ministry of Agriculture of the applicant and the Animal Husbandry and Veterinary Department of Hubei Academy of Agricultural Sciences were selected. The 142 Large White pigs in the research institute were used as test materials. The AluI-RFLP method established in Example 2 was used for polymorphism detection, and the correlation between different genotypes of pig AluI-RFLP and pig litter size traits was analyzed. The SAS statistical software (SAS Institute Inc, Version 8.0) GLM program was used for single-marker analysis of variance, and the REG program was used to calculate the additive effect and dominant effect of the gene, and the signifi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com