DNA chip based cipher system
A DNA chip and encryption system technology, applied in the field of DNA chip-based encryption system, can solve the problems of high operation cost, high cost, and error-prone decryption, and achieve the effect of clear information transmission, cost saving, and cost increase
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
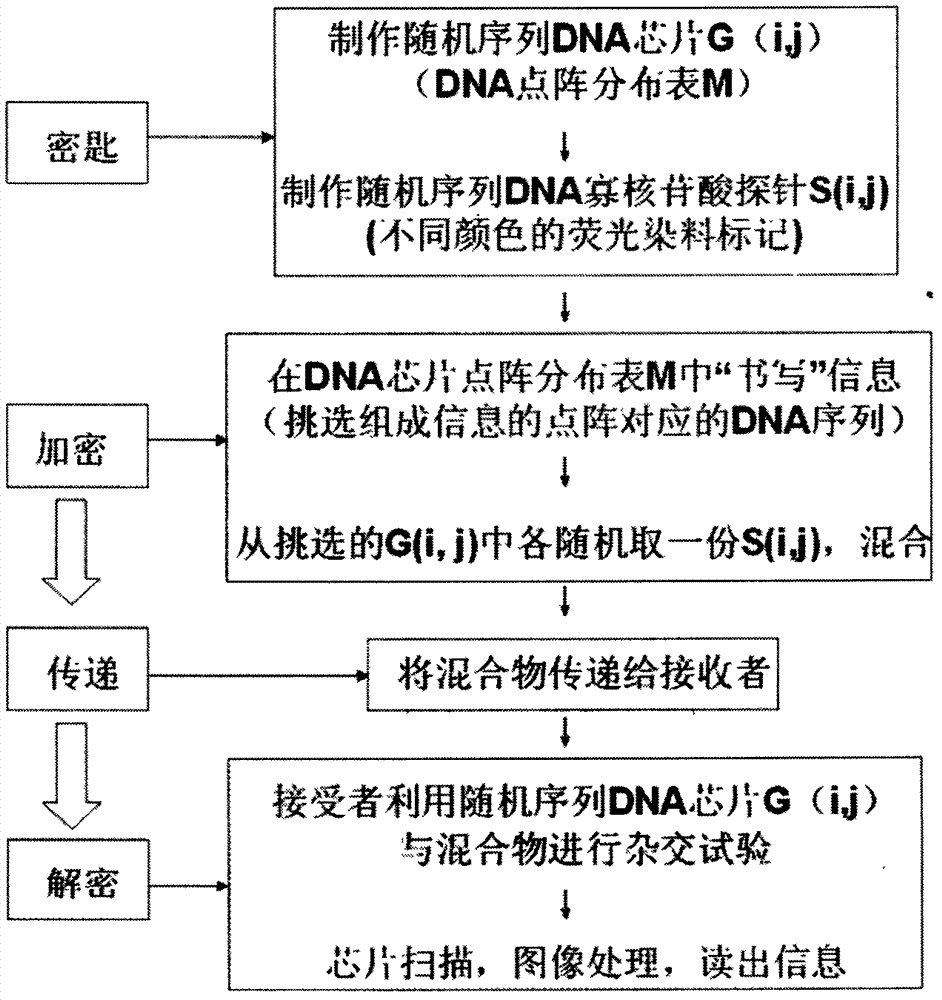
[0023] The making of embodiment 1 random sequence DNA chip
[0024] First, a random DNA sequence is obtained. A random sequence composed of 0 and 1 numbers can be generated by a random sequence generator, and then converted into a random DNA sequence composed of "ATGC" according to "00=A, 01=T, 10=G, 11=C".
[0025] Then, the random DNA sequences generated above are screened. DNA sequences with GC content greater than 60% and less than 40% and DNA sequences with the same letters of A, T, G, and C or AT and GC repeated more than 8 times were excluded.
[0026] Thirdly, send the DNA sequence of about 1-2 kb obtained above to a DNA synthesizer to artificially synthesize DNA fragments.
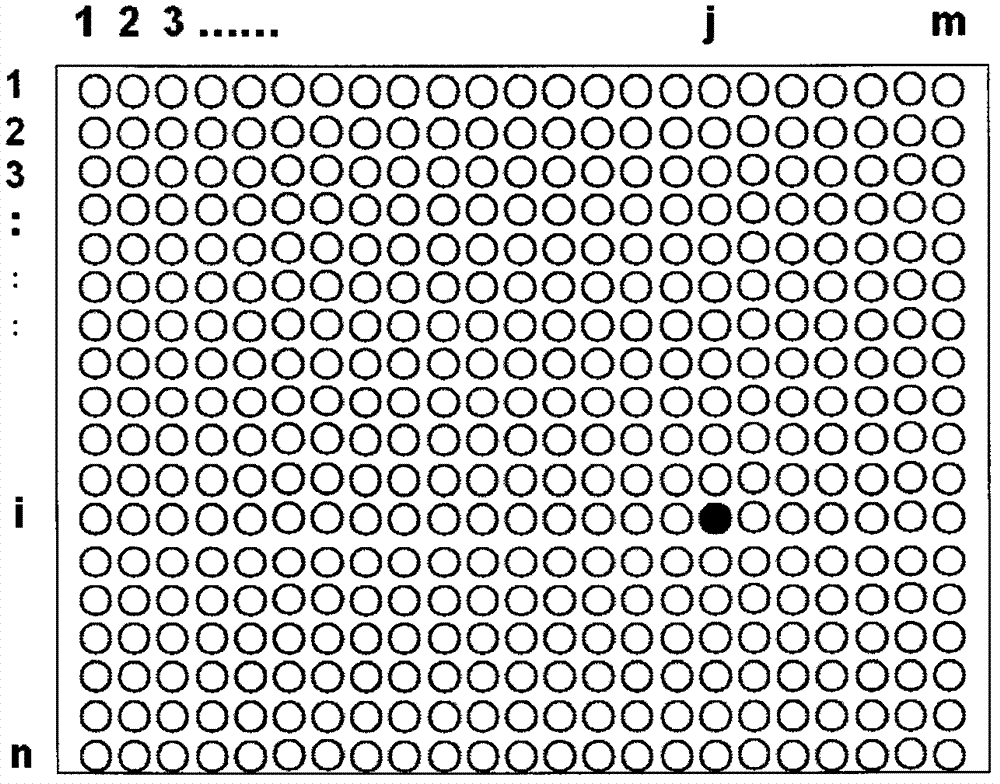
[0027] Fourth, make a DNA chip dot matrix distribution map M, and compile the DNA sequence fragment name and sequence (G(i, j)) corresponding to each dot matrix in Excel file format.
[0028] Finally, through the DNA chip production system, the random sequence DNA fragments were microprinted on...
Embodiment 2
[0029] Embodiment 2 makes random sequence DNA oligonucleotide probe
[0030] First, for each random DNA sequence G(i, j), randomly intercept a 15bp long DNA sequence S(i, j), and use the BLAST program to analyze all DNA sequences G(i, j) on the random sequence DNA chip Search and compare, if it is found that the selected S(i, j) is the only one with 100% homology and less than 40% homology with other sequences, then the S(i, j) can be used to synthesize DNA oligonuclei nucleotide probe. Each G(i, j) can select multiple S(i, j) for synthesizing multiple copies of oligonucleotide probes.
[0031] Second, the selected random DNA sequence fragment S(i, j) is synthesized with a DNA synthesizer using its reverse complementary sequence, and each S(i, j) is labeled with a different fluorescent dye, such as Cy3 or Cy5-labeled.
[0032] Third, the synthesized oligonucleotide probes were purified by PAGE and stored in a centrifuge tube in powder form. The amount of synthesis can be a...
Embodiment 3
[0034] Embodiment 3 information encryption method
[0035] First, the processing of plaintext information. According to the agreement of the communication parties, the plaintext information can undergo various transformations, such as conversion into binary form, Braille code or Morse code form, etc. It is also possible not to perform conversion.
[0036]Then, the above-mentioned processed plaintext information is marked on the DNA chip lattice distribution map M in the form of a 2-dimensional lattice. The process is similar to "writing" on the dot matrix map M of the DNA chip, "writing" the above-mentioned plaintext information on the dot matrix map M of the DNA chip.
[0037] Third, the above-mentioned selected and labeled lattices correspond to the corresponding random DNA sequences G(i, j). According to its marked color and corresponding G(i, j), the corresponding oligonucleotide probe S(i, j) is taken out from the synthesized oligonucleotide probe library. Since each ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com