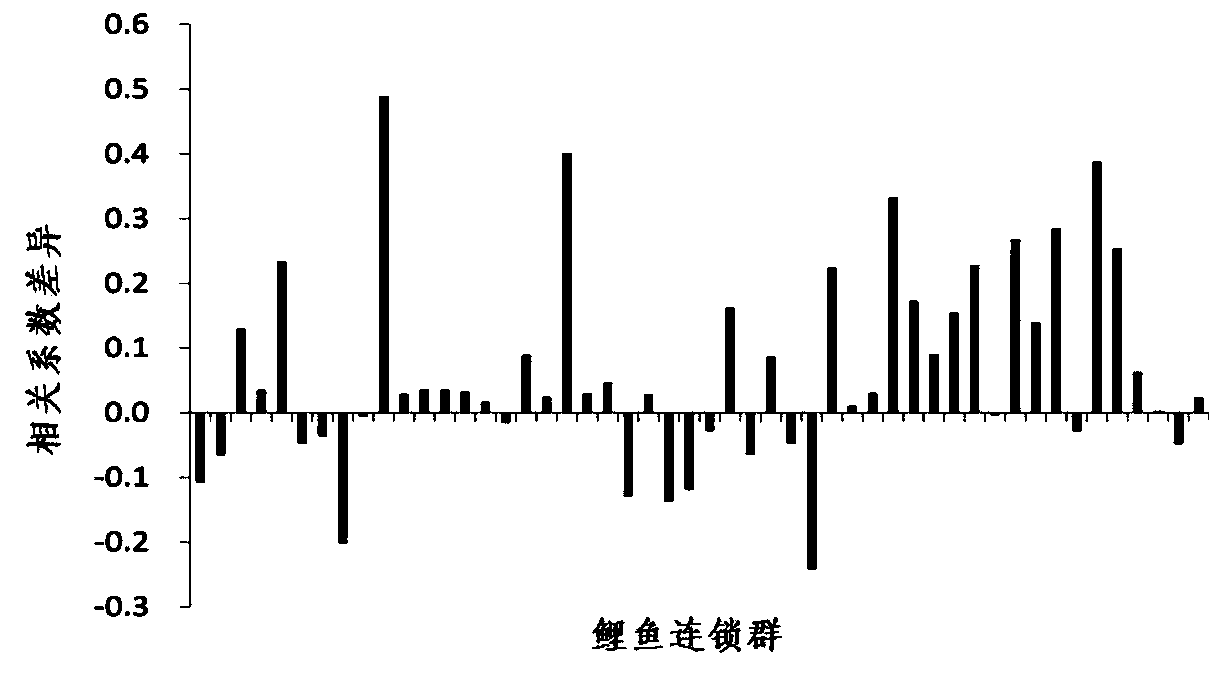
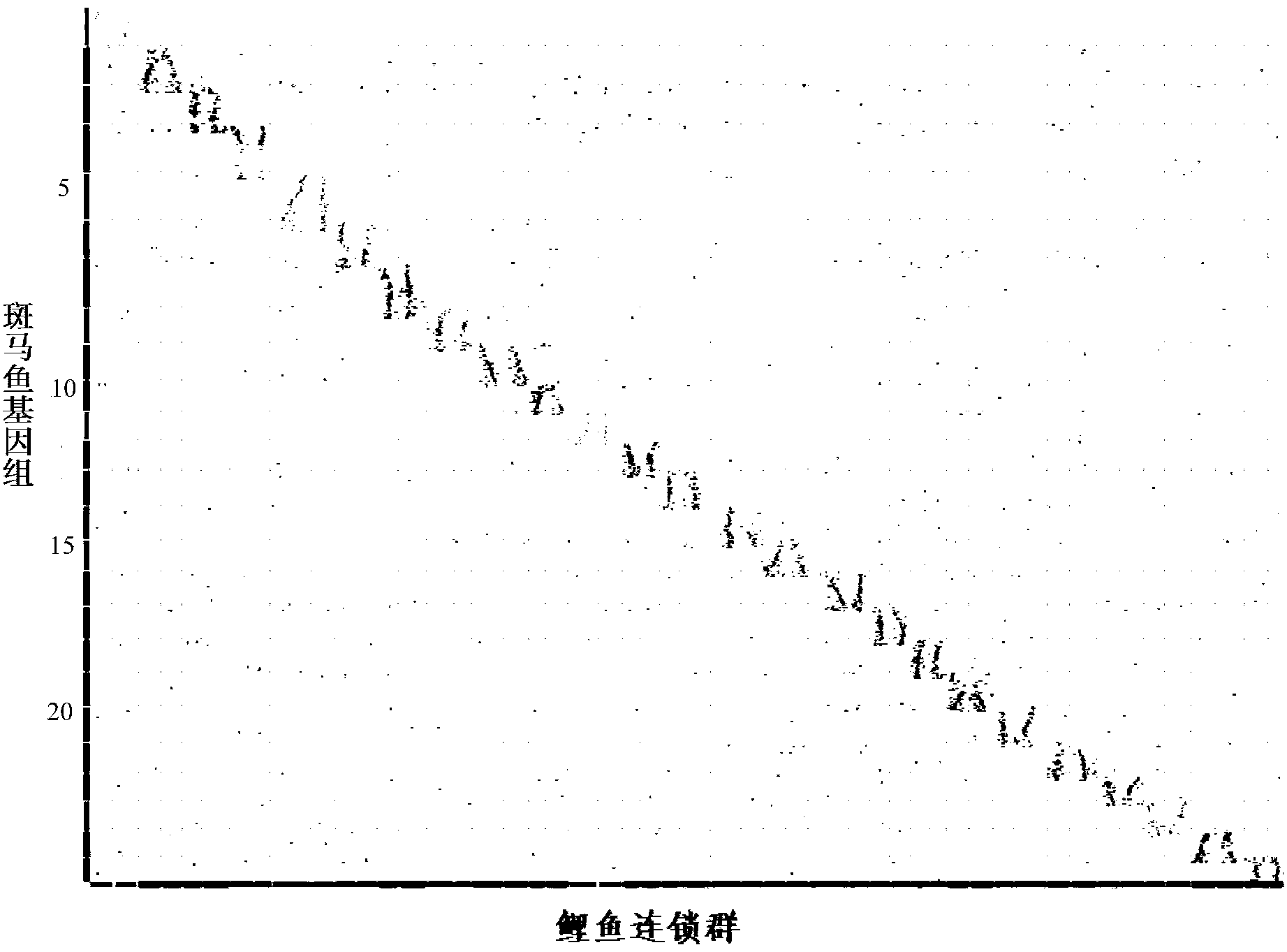
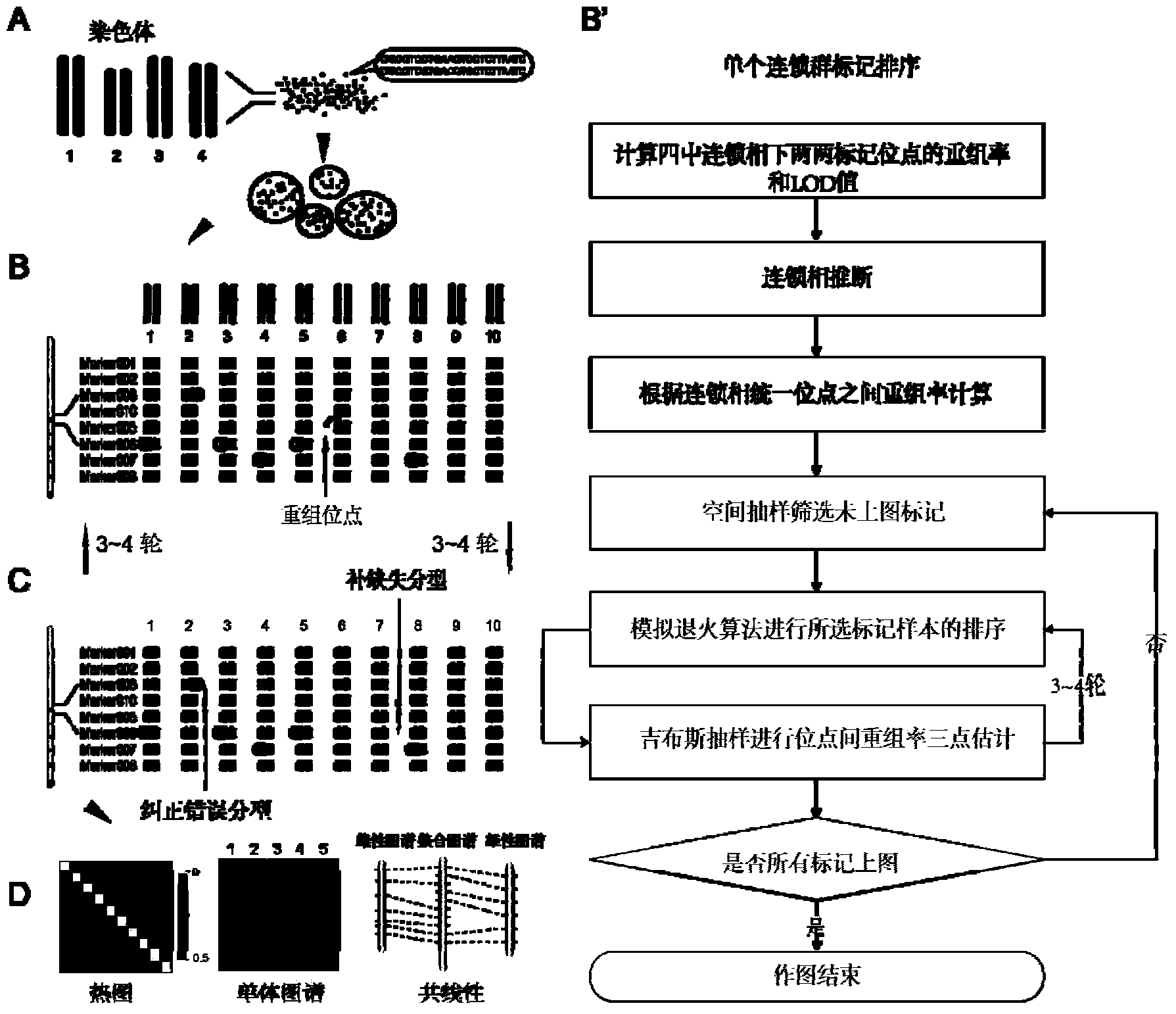
Construction and evaluation of parting High Map on basis of high throughput
A genetic map, high-throughput technology, applied in biochemical equipment and methods, microbial measurement/inspection, instruments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1: Application of HighMap to the construction of genetic map of F1 population of 224 carp strains
[0071] Carp genetic map construction, including the following steps:
[0072] 1. Selection of carp colony (purchased from Heilongjiang Fisheries Research Institute): the colony is F1 colony with 224 strains and 2 parents. The high-throughput sequencing instrument is Illumina GA IIx.
[0073] 2. Use SLAFseq technology (the method of patent CN103088120A) to develop and type the whole genome markers of carp populations, and perform data identification processing on the original sequencing data. After cluster analysis and error correction, 185,014 SLAF tags were obtained; as shown in Table 1 Shown as the number of SLAF tags and sequencing depth statistics.
[0074] Table 1 Statistics on the number of total sequencing tags and sequencing depth
[0075]
[0076] 3. As shown in Table 2, the marker coding rules for the F1 population were selected. The above-mentione...
Embodiment 2
[0092] Example 2 HighMap is used to simulate genetic map construction and map quality assessment of simulating typing data
[0093] Genetic map construction and map quality assessment based on simulation data, including the following steps:
[0094] 1. Use the Monte Carlo method to simulate the missing typing and errors that may be caused by different sequencing depths. The results show that using the sequencing method for genotyping, the missing typing and typing errors related to the sequencing depth are inevitable. For high heterozygous In terms of species, the problem of typing quality caused by sequencing depth is particularly prominent. Table 6 shows the proportion of typing errors and typing deletions that may occur when sequencing and typing sites with different separation patterns at different sequencing depths. When the average sequencing depth is 1x, the sites with abxcd separation patterns, The typing error was as high as 34%. As the sequencing depth gradually inc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com