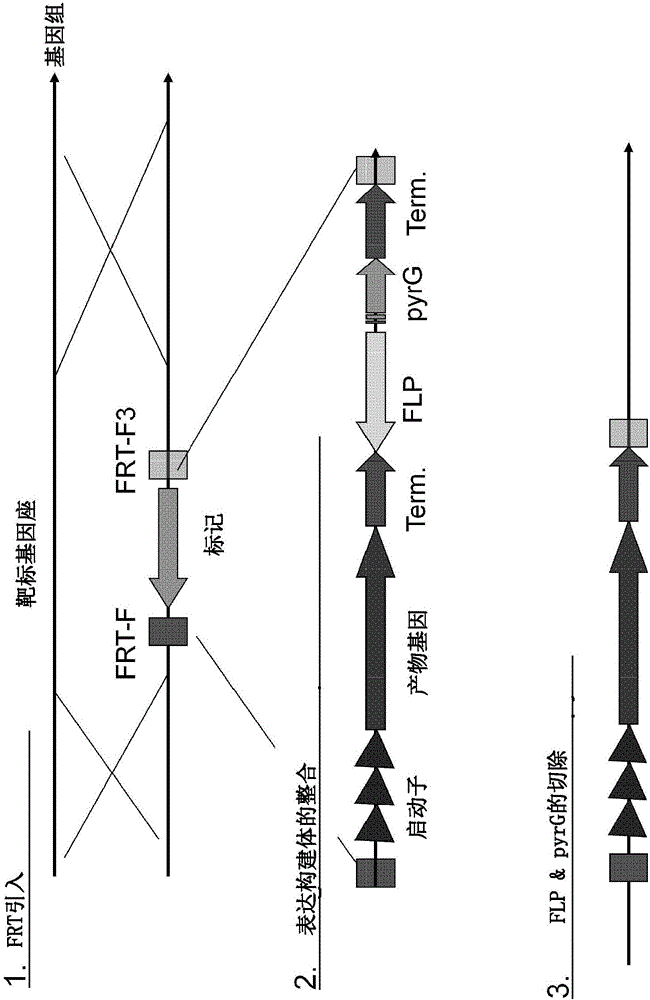
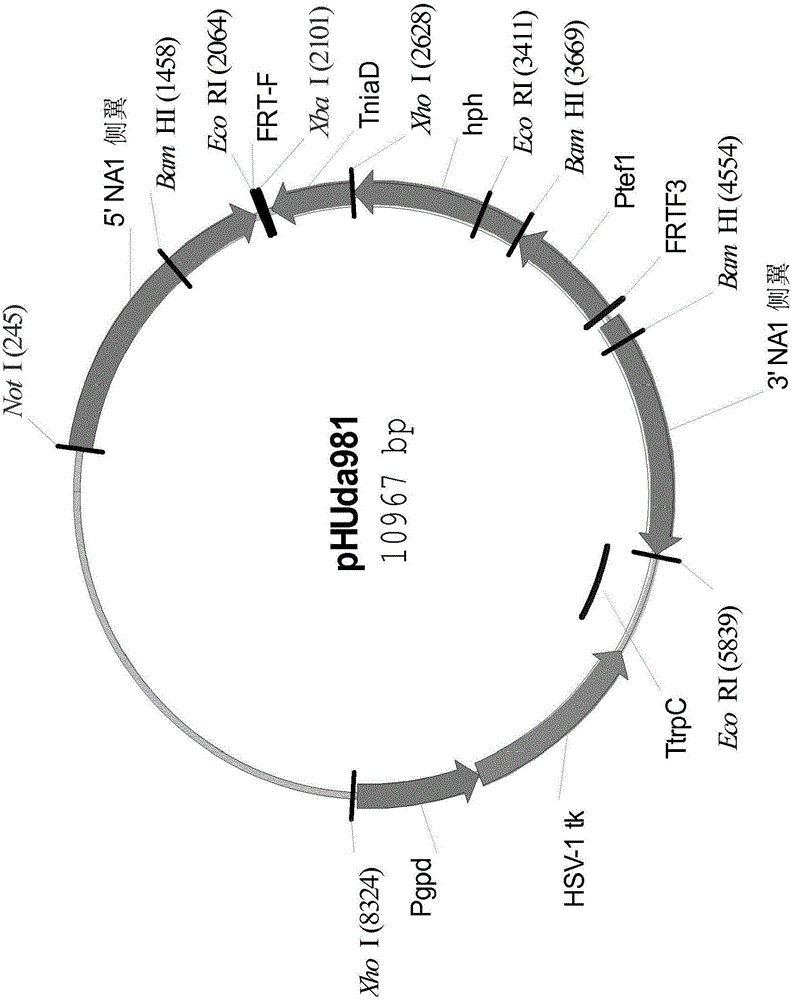
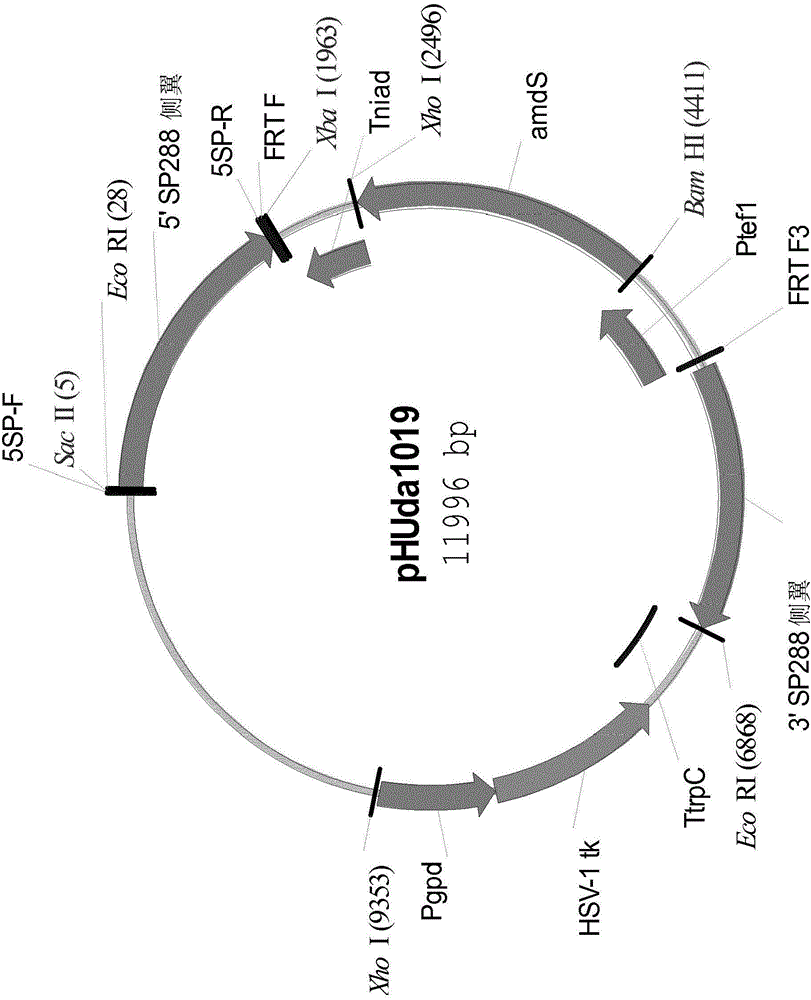
Simultaneous site-specific integrations of multiple gene-copies in filamentous fungi
A site-specific, fungal technology that can be used in genetic engineering, other methods of inserting foreign genetic material, stably introducing foreign DNA into chromosomes, etc., and can solve problems such as irreversible excision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 10
[0206] Plasmid pJaL504 is described in Example 10.
[0207] Plasmid pJaL504-delta-Bglll is described in Example 10.
[0208] Plasmid pJaL554 is described in Example 1 of patent WO2000 / 050567A1.
[0209] Plasmid pJaL574 is described in Example 10.
[0210] Plasmid pJaL835 is described in Example 10.
[0211] Plasmid pJaL955 is described in Example 10.
[0212] Plasmid pJaL1022 is described in Example 10.
[0213] Plasmid pJaL1025 is described in Example 10.
[0214] Plasmid pJaL1027 is described in Example 10.
[0215] Plasmid pJaL1029 is described in Example 10.
[0216] Plasmid pJaL1120 is described in Example 10.
[0217] Plasmid pJaL1123 is described in Example 10.
[0218] Plasmid pJaL1183 is described in Example 10.
[0219] Plasmid pJaL1194 is described in Example 10.
[0220] Plasmid pJaL1202 is described in Example 10.
[0221] Plasmid pToC65 is described in patent WO91 / 17243.
[0222] Plasmid pUC19: This construction is described in Vieira et al., 1982, Ge...
Embodiment 1
[0254] Example 1. Introduction at the neutral amylase I (NAI) locus of Aspergillus niger NN059095 FRT site
[0255] Construction of hygromycin B resistance gene expression plasmid pHUda966
[0256] Based on the nucleotide sequence information in GENBANK (ID#AB007770), the following primers Tef-F and Tef-R were designed to introduce EcoRI / SpeI and BamHI sites, respectively, to isolate the promoter region of Aspergillus oryzae tef1 (translation elongation factor 1 / Ptef1):
[0257] Tef-F (SEQ ID NO: 1): gaattcactagtggggttcaaatgcaaacaa
[0258] Tef-R (SEQ ID NO: 2): ggatcctggtgcgaactttgtagtt
[0259] A PCR reaction was performed using the primer pair Tef-F and Tef-R with the genomic DNA of Aspergillus oryzae strain BECh2 as a template. The reaction products were separated on 1.0% agarose gel, and the 0.7kb product band was excised from the gel. The 0.7 kb amplified DNA fragment was digested with BamHI and EcoRI and ligated into the Aspergillus expression cassette pHUda4...
Embodiment 2
[0289] Example 2. Introduction of an FRT site at the acid-stable amylase locus of Aspergillus niger NN059095 point
[0290] Construct the expression plasmid pHUda976 of Aspergillus nidulans acetamidase gene (amdS).
[0291] Based on the nucleotide sequence information in EMBL:AF348620, the following primers amdS-F and amdS-R were designed to introduce the BamHI and XhoI sites, respectively, to isolate the coding region of the amdS gene:
[0292] amdS-F (SEQ ID NO: 19): ggatccaccatgcctcaatcctgg
[0293] amdS-R (SEQ ID NO: 20): ctcgagctatggagtcaccacatttcccag
[0294] A PCR reaction was performed using the primer pair amdS-F and amdS-R with the genomic DNA of Aspergillus nidulans strain NRRL1092 as template. The reaction products were separated on a 1.0% agarose gel, and a 1.0 kb product band was excised from the gel. The 1.9 kb amplified DNA fragment was digested with BamHI and XhoI and ligated into the Aspergillus expression cassette pHUda440-Ptef-Tnia digested with Ba...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com