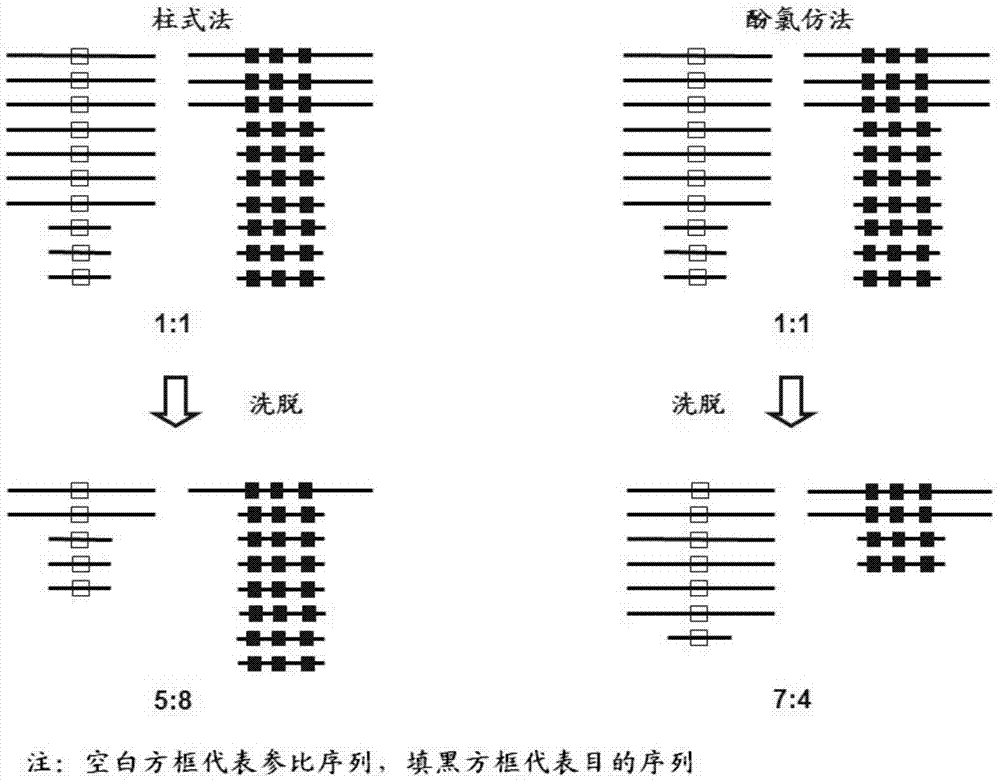
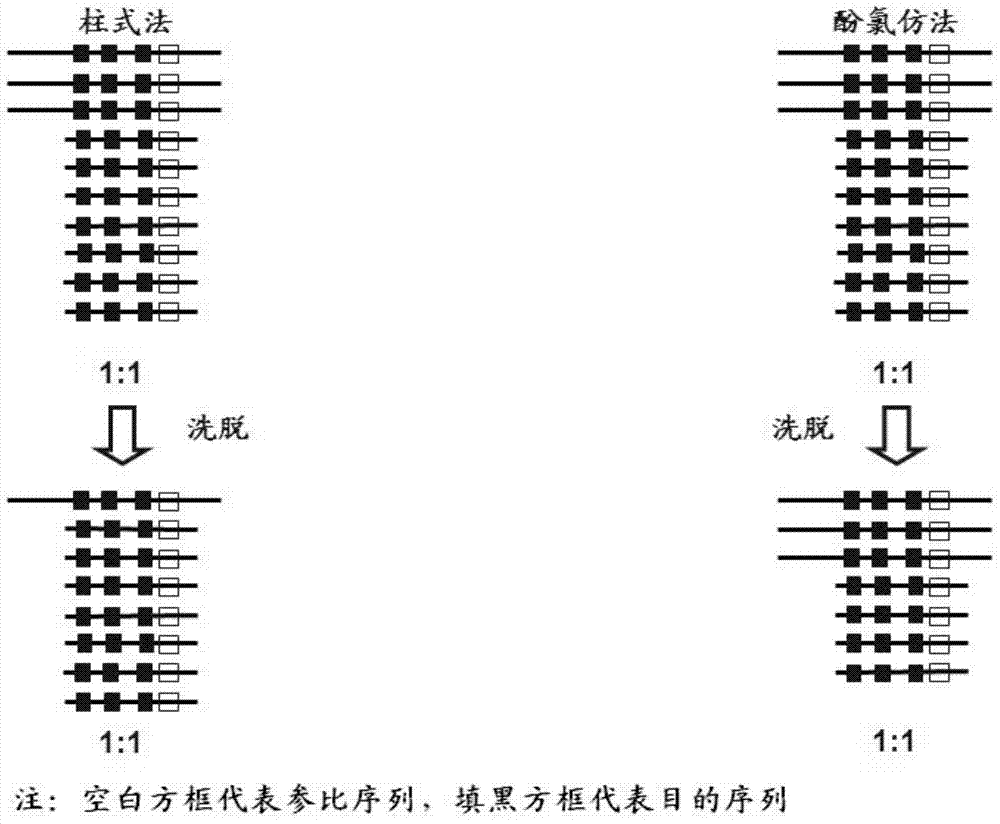
Fluorescent quantitative PCR (Polymerase Chain Reaction) kit for diagnosing human spinal muscular atrophy
A spinal muscular atrophy, fluorescence quantitative technology, applied in the field of biochemistry, can solve problems such as high cost, heavy burden on patients, difficulty in large-scale population screening and technology promotion
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0071] Example 1 (Analysis of Fluorescence Quantitative Detection Result by Ordinary Reference Gene Method)
[0072] 1. Reagent composition:
[0073] (1) Among the pair of shared primers for specifically amplifying a certain sequence of SMN1 and SMN2 genes,
[0074] The upstream primer sequence is: 5'-CATCCATATAAAGCTATCTATATAG-3' (SEQ NO.1),
[0075] The downstream primer sequence is: 5'-CTTAATTTAAGGAATGTGAGCACCT-3' (SEQ NO.2);
[0076] (2) Among the pair of primers for specifically amplifying the NAIP gene,
[0077] The upstream primer sequence is: 5'-TTGTGACTTATGAGCCGTACAGC-3' (SEQ NO.3),
[0078] The downstream primer sequence is: 5'-AGGCTACAGCAGAAGCACTGAAT-3' (SEQ NO.4);
[0079] (3) In the pair of primers for specifically amplifying a certain sequence of the reference sequence GAPDH gene,
[0080] The upstream primer sequence is: 5'-CAGGAGTGAGTGGAAGACAGAAT-3' (SEQ NO.11),
[0081] The downstream primer sequence is: 5'-GACCATATTGAGGGACACAAGGT-3' (SEQ NO.12);
[0082...
Embodiment 2
[0108] Example 2 (Analysis of Fluorescence Quantitative Detection Result by Proximity to Reference Sequence Method)
[0109] 1. Reagent composition:
[0110] (1) Among the pair of shared primers for specifically amplifying SMN1 and SMN2 genes,
[0111] The upstream primer sequence is: 5'-CATCCATATAAAGCTATCTATATAG-3' (SEQ NO.1),
[0112] The downstream primer sequence is: 5'-CTTAATTTAAGGAATGTGAGCACCT-3' (SEQ NO.2);
[0113] (2) Among the pair of primers for specifically amplifying the NAIP gene,
[0114] The upstream primer sequence is: 5'-TTGTGACTTATGAGCCGTACAGC-3' (SEQ NO.3),
[0115] The downstream primer sequence is: 5'-AGGCTACAGCAGAAGCACTGAAT-3' (SEQ NO.4);
[0116] (3) Among the pair of primers for specifically amplifying the reference sequence,
[0117] The upstream primer sequence is: 5'-GAACACACATGTCAGAAGTCTAAG-3' (SEQ NO.5),
[0118] The downstream primer sequence is: 5'-ACCTCCAGTTAGATCTTCACTTCT-3' (SEQ NO.6);
[0119] (4) The fluorescent probe for specifically...
Embodiment 3
[0145] Example 3 (Sensitivity and Accuracy Small Sample Evaluation of the Kit)
[0146] 1. The composition of the kit:
[0147] (1) Among the pair of shared primers for specifically amplifying a certain sequence of SMN1 and SMN2 genes,
[0148] The upstream primer sequence is: 5'-CATCCATATAAAGCTATCTATATAG-3' (SEQ NO.1),
[0149] The downstream primer sequence is: 5'-CTTAATTTAAGGAATGTGAGCACCT-3' (SEQ NO.2);
[0150] (2) Among the pair of primers for specifically amplifying a certain sequence of the NAIP gene,
[0151] The upstream primer sequence is: 5'-TTGTGACTTATGAGCCGTACAGC-3' (SEQ NO.3),
[0152] The downstream primer sequence is: 5'-AGGCTACAGCAGAAGCACTGAAT-3' (SEQ NO.4);
[0153] (3) In the pair of primers for specifically amplifying a certain segment of the reference sequence,
[0154] The upstream primer sequence is: 5'-GAACACACATGTCAGAAGTCTAAG-3' (SEQ NO.5),
[0155] The downstream primer sequence is: 5'-ACCTCCAGTTAGATCTTCACTTCT-3' (SEQ NO.6);
[0156] (4) The fl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com