Luciferase complementary image detection method and carrier kit for detecting protein interaction
A luciferase and image detection technology, applied in the field of protein research, can solve problems such as gateways that have not yet been seen
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
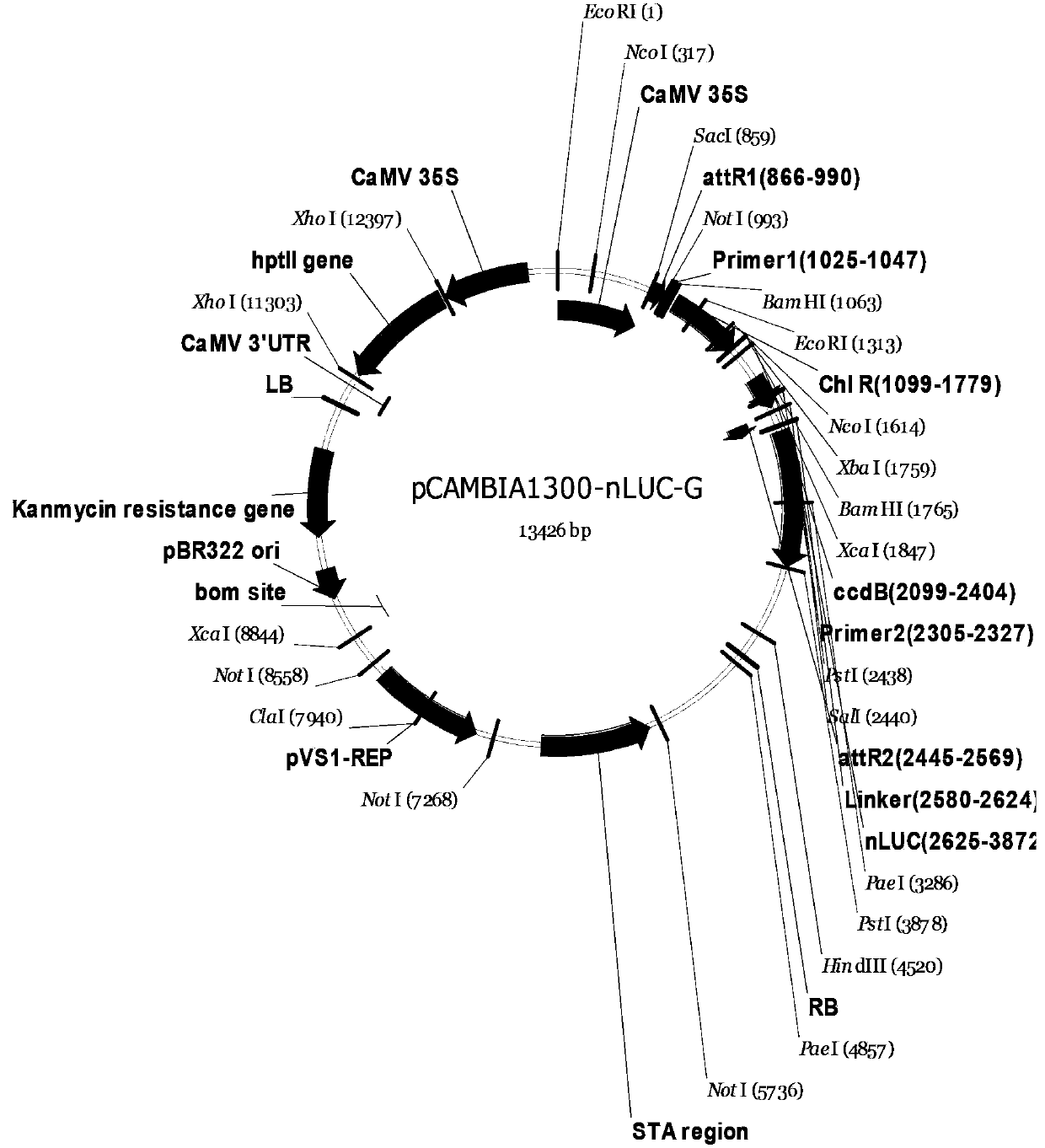
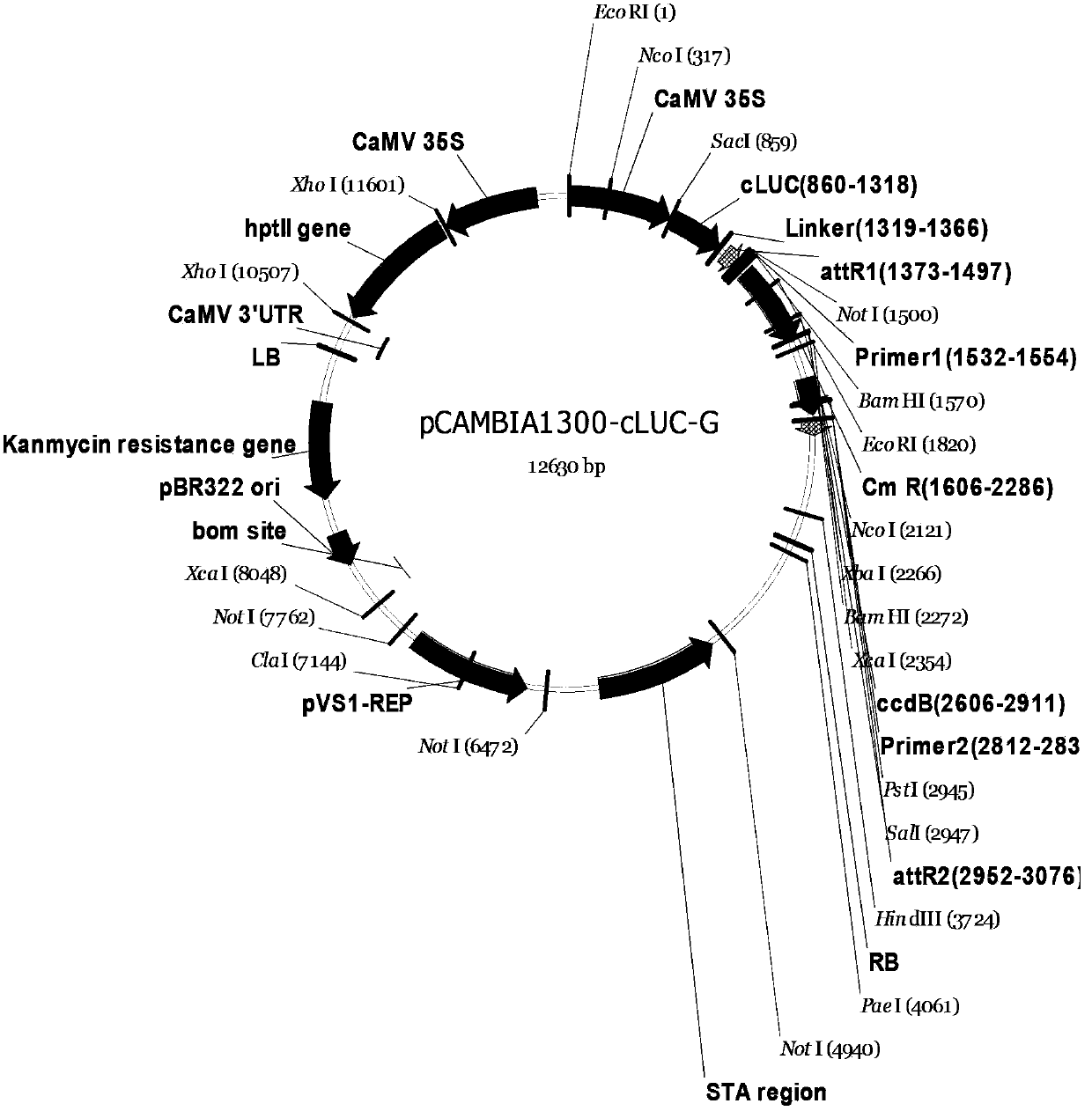
[0104] Example 1. Construction of LCI technology Gateway vector: (Nluc is located at the C-terminus of the recombination site)
[0105] 1.1.pNG ( figure 1 ) build
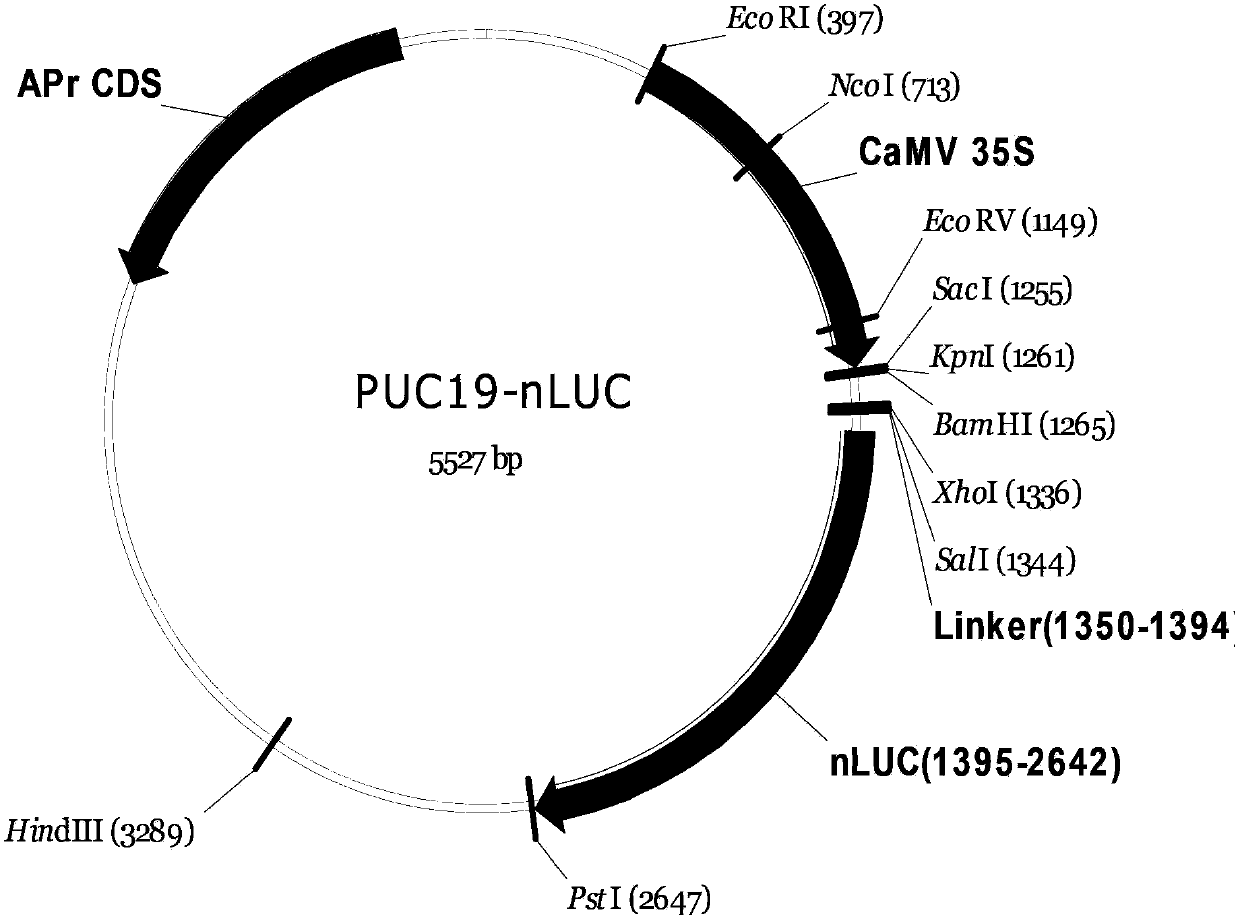
[0106] Step 1. Digest pUC-Nluc (i.e. 35S::Nluc) with restriction enzyme KpnI-SalI ( image 3 ), excise the multiple cloning site sequence, and smooth the end of the obtained fragment to facilitate insertion of the RFC2.1 recombinant fragment.
[0107] Step 2. Use T4DNApolymerase (NEB) to simultaneously cut the 3'-overhanging end of the verified fragment in step 1 (KpnI forms the end), and fill in the 5'-overhanging end (SalI forms the end); then use alkaline Phosphatase dephosphorylates its terminus to prevent self-ligation in the ligation reaction to obtain pUC-Nluc-KS.
[0108] Step 3. Cloning of RFC2.1 recombination site into pUC-Nluc-KS: Ligate pUC-Nluc-KS with RFC2.1 fragment (see Invitrogen), and transform the ligation product into One ccdB Survival TM 2T1 R Competent cells (Invitrogen, resistant to cc...
Embodiment 2
[0121] Example 2. The LCI technology Gateway vector is used to detect the interaction between protein genes AtSGT1a and AtRAR1 to construct a fusion vector and introduce it into Agrobacterium GV3101 or GV2260 (both are acceptable).
[0122] Objective: To detect the interaction between AtSGT1a and AtRAR1, and use the interaction between AtSGT1a and AtR77 as a negative control. AtR77 comprises only the N-terminal 77 amino acids of AtRAR1.
[0123] The original LCI expression vectors pCAMBIA-Nluc and pCAMBIA-Cluc were used as controls to verify the applicability of pNG and pCG expression vectors in LCI technology.
[0124] Step 1. Construct the entry vector of the target gene (see invitrogen Catalog nos.12535-019 manual for details)
[0125] The construction process of the entry vector of AtSGT1a gene, AtRAR1 gene and AtR77 gene:
[0126] (1) PCR amplification of target gene with attB recombination site
[0127] By using primers with attB recombination sites to amplify the gen...
Embodiment 3
[0180] Example 3. Operational steps for high-throughput screening of protein interactions.
[0181] Step 1. cDNA library construction (see Invitrogen Ca.no.18248-013 "Super Plasmid System with Technology for cDNA Synthesis and Cloning"):
[0182] (In the pNG and pCG vectors, there are BamHI and SalI restriction sites adjacent to the attR1 and attR2 recombination sites, and there are no restriction sites outside attR1-attR2 on the vector, so the BamHI and SalI restriction sites are suitable as To construct the linker of cDNA library, see Figure 16 .
[0183] Specific steps are as follows:
[0184] Extract the mRNA of the sample; synthesize the first strand of cDNA; use poly(T)+BamHI digestion recognition site (ggatcc) as the reverse transcription primer;
[0185] Synthesize the second strand of cDNA; add SalI linker (gtcgac); obtain double-stranded cDNA with BamHI and SalI restriction sites at both ends.
[0186] BamHI-SalI digestion of double-stranded cDNA, and vector...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com