Application of SATB1 to treatment of cutaneous T cell lymphomata
A lymphoma, cell technology, applied in gene therapy, medical preparations containing active ingredients, antitumor drugs, etc., can solve the problem of unclear role of SATB1
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
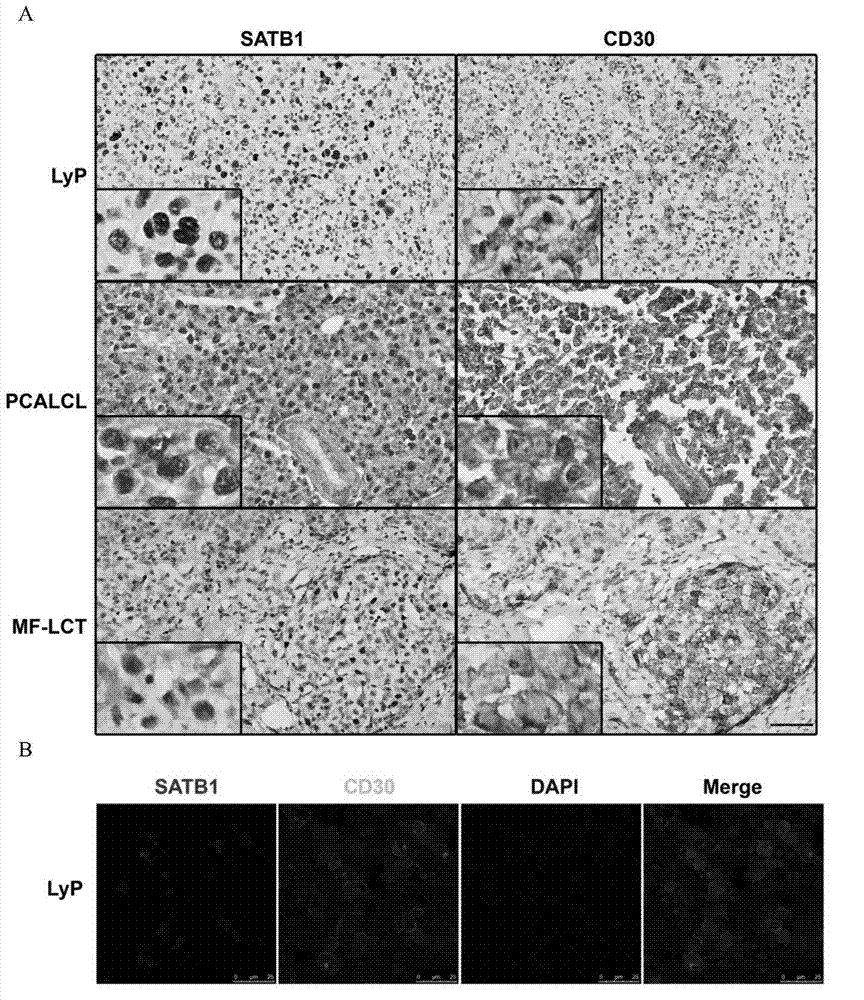
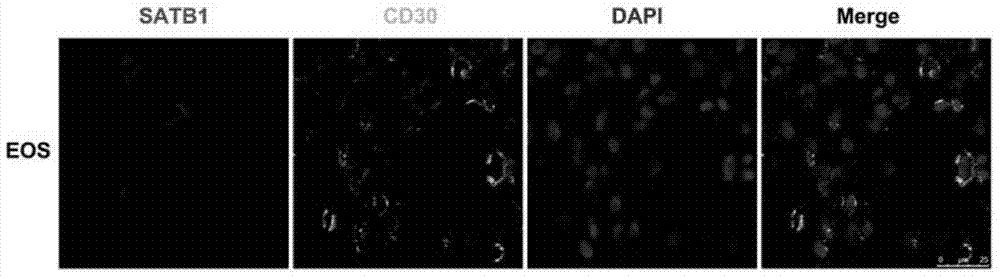
[0074] Example 1. SATB1 is specifically highly expressed in CD30+ neoplastic T cells of cutaneous T-cell lymphoma
[0075] 1. Immunohistochemical staining
[0076] Immunohistochemical staining was performed on paraffin specimens of MF large cell transformation (MF-LCT), lymphomatoid papulosis (LyP), and primary cutaneous anaplastic large cell lymphoma (PCALCL), as follows:
[0077] (1) Embed the specimen in paraffin, make 5 μm slices, routinely dewax into water, put in xylene I, II for 10 minutes each, absolute ethanol I, II, ethanol aqueous solution I, 95% by volume II, 80% ethanol water solution by volume for 5 minutes each, fully hydrated with distilled water for 3-5 minutes, and rinsed with PBS (pH7.4) 3 times, 5 minutes each time.
[0078] (2) Antigen retrieval: Put the tissue slices into 0.01mol sodium citrate buffer, heat in a microwave oven on high heat until boiling, then switch to medium heat and heat for 10 minutes, take it out and let it stand to cool to room temp...
Embodiment 2
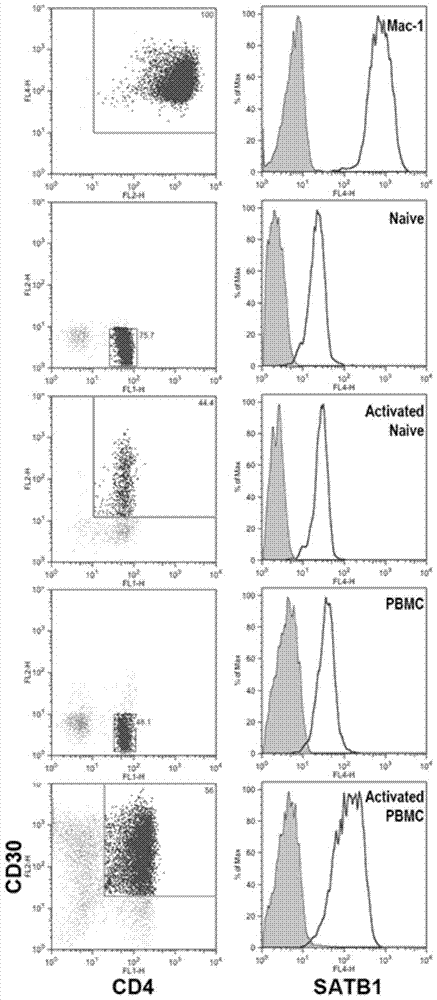
[0141] Example 2. The expression of SATB1 increases with the progression of cutaneous T-cell lymphoma (CTCL)
[0142] The sources and characteristics of the following cell lines are shown in Table 2.
[0143] Table 2 Sources and characteristics of the cell lines used
[0144]
[0145] The cell culture used in the embodiment adopts RPMI1640 liquid culture medium, adds the fetal calf serum that volume percentage is 10%, 100U / ml penicillin, 0.1mg / ml streptomycin, and the cell is the suspension type growth in the culture medium, inserts Constant temperature 37°C, humidified 5% CO 2 Cell incubator, according to the state of the cells, replace the medium for passage.
[0146] 1. Extract the total RNA of Hut78, Jurkat, Mac-1, Mac-2A and Mac-2B and synthesize the first strand of cDNA.
[0147] 2. Real-time fluorescent quantitative PCR
[0148] Using the cDNA obtained in step 1 as a template, PCR amplification was performed with specific primers for the SATB1 gene, and the produ...
Embodiment 3
[0195] Example 3. The high expression of SATB1 promotes the malignant biological phenotype of cutaneous T-cell lymphoma disease
[0196] 1. Lentiviral vector
[0197] The GV118 lentiviral vector is a cloning vector, and its structure is as follows: Figure 6 shown.
[0198] Figure 6 Among them, the full length of the GV118 vector is 10307bp, and HpaI and XhoI are multiple cloning sites. GFP was used as a fluorescent marker for the selection of transfected cells. The ampicillin resistance gene can be used as a screening gene for cell clones.
[0199] 2. Construction of RNA interference lentiviral vector
[0200] (1) Design of shRNA sequence:
[0201] For the SATB1 gene sequence, use the RNA interference sequence design principles provided in the public website to design the following RNA (shRNA) sequence interference target sequence:
[0202] shRNA0:
[0203] 5'-GCGTTAACCGATTCGGTGCTACTCTATGCAAGAGATCGAGTAGCACCGAATCGCTCGAG-3'; (SEQ ID No. 7)
[0204] shRNA1:
[0205] 5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com