Quantitative detection method of antibiotics resistance gene in soil
A quantitative detection method and antibiotic resistance technology, applied in the detection field of soil environmental pollutants, can solve the problems of no resistance gene and low sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
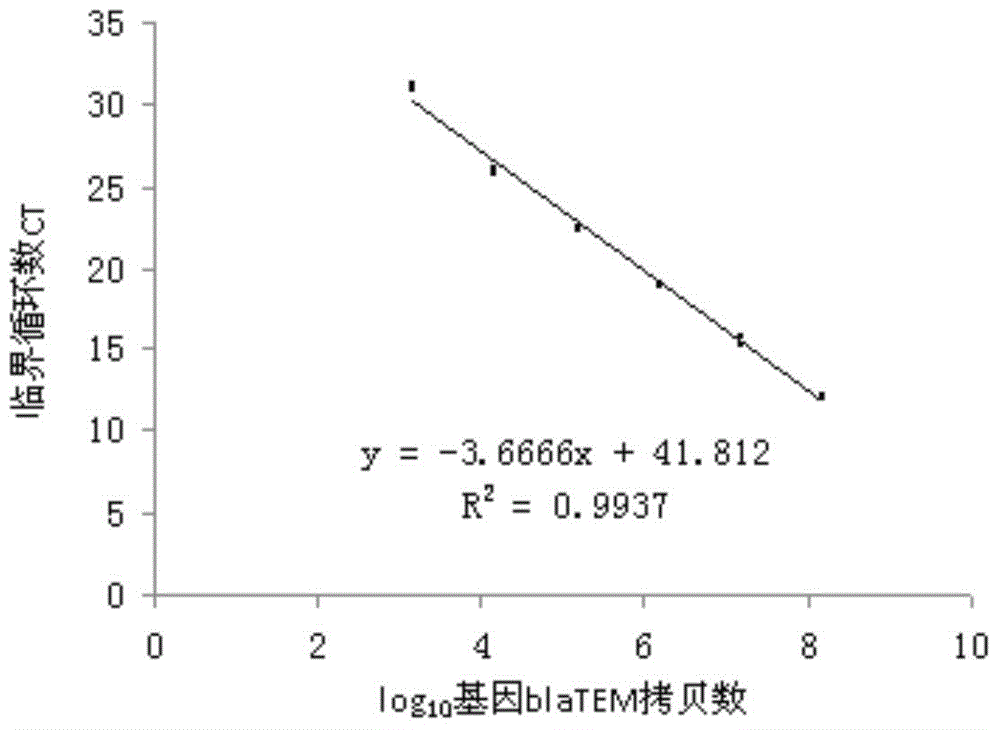
[0049] Example 1, Quantitative detection of penicillin resistance gene blaTEM in different soils
[0050] The β-lactamase encoded by blaTEM can invalidate penicillin. Get fresh forest soil, pasture soil, and tea garden soil to measure the soil water content. Each soil sample and pig manure sample weigh 0.25g to extract DNA, and the volume is 100 μ L [the pig manure The feces produced by pigs taking antibiotics for a long time; the DNA of the above soil samples and pig manure samples were extracted using the Fast DNA spin kit for soil].
[0051] 30 known blaTEM gene sequences (GenBank database) were selected for comparison, and it was found that the blaTEM gene was well conserved. The primers were redesigned according to the conserved sequences, and the generated target fragment was 129 bp. This pair of primers can be considered as universal primers for detecting blaTEM gene.
[0052] Use pig feces sample DNA as a template for PCR. The common PCR reaction system is a 20 μL rea...
Embodiment 2
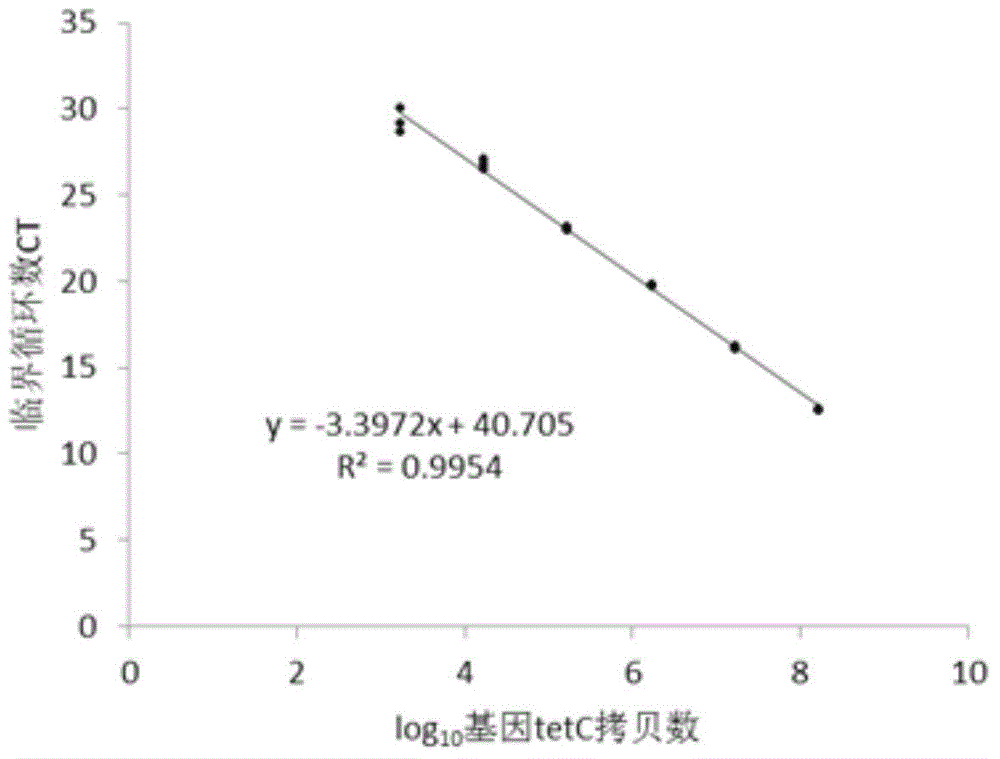
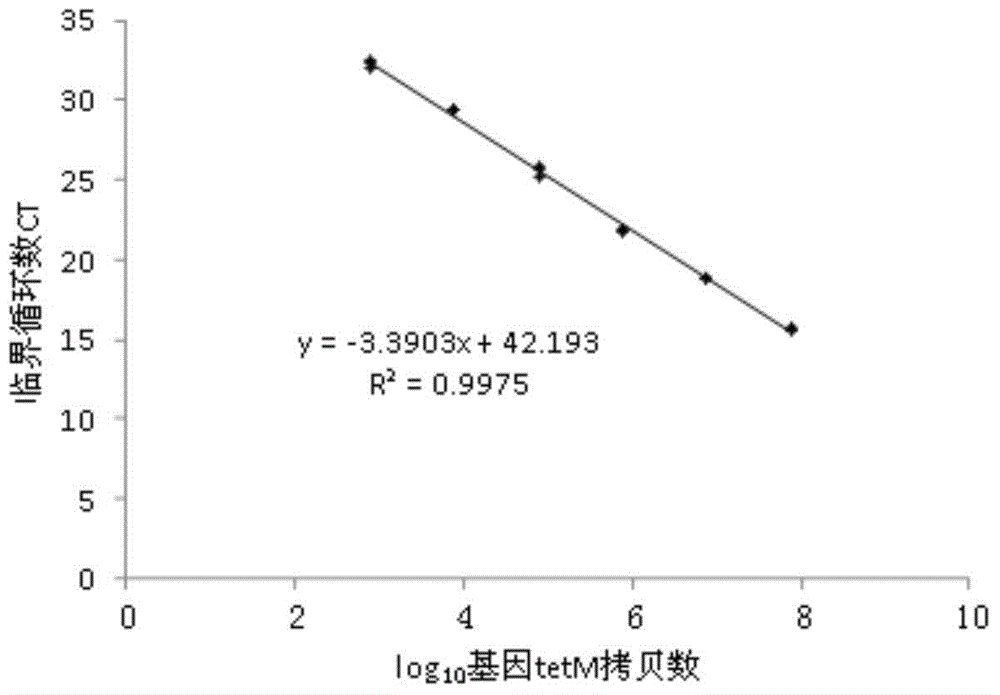
[0060] Example 2, Quantitative comparison of tetracycline resistance genes tetC and tetM in different soils
[0061] tetC encodes an efflux pump protein, which can excrete tetracycline out of the cell, thereby reducing the intracellular drug concentration to play a protective role. tetM encodes a ribosome protection protein, which causes a change in ribosome configuration, preventing tetracycline from binding to ribosomes and failing.
[0062] Get fresh forest soil, pasture soil, tea garden soil and measure soil water content, each soil sample and pig manure sample weigh 0.25g to extract DNA, and volume is 100 μ L [the described pig manure is the excrement that the pig place that takes antibiotic for a long time; The above-mentioned soil The DNA of the soil samples and pig manure samples were extracted with the Fast DNA spin kit for soil].
[0063] The primer sequences before and after tetC are GCGGGATATCGTCCATTCCG, GCGTAGAGGATCCACAGGACG, respectively, and the target fragment ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| PCR efficiency | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
| PCR efficiency | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com