AHAS mutants
A technology of variants and expression vectors, applied in the field of tolerance compositions, can solve problems such as not describing imidazolinone-specific resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0150] 1. Dilute the product for foliar application with water. For seed treatment purposes, such products may be applied to the seed with or without dilution.
[0151] A) Water-soluble concentrate (SL, LS)
[0152] 10 parts by weight of the AHAS-inhibiting herbicide is dissolved in 90 parts by weight of water or a water-soluble solvent. Alternatively, wetting agents or other auxiliaries are added. After dilution with water, the AHAS-inhibiting herbicide was dissolved, thus obtaining a formulation containing 10% (w / w) of the AHAS-inhibiting herbicide.
[0153] B) Dispersible Concentrate (DC)
[0154] 20 parts by weight of an AHAS-inhibiting herbicide was dissolved in 70 parts by weight of cyclohexanone, and 10 parts by weight of a dispersant (for example, polyvinylpyrrolidone) was added. Dilution with water produces a dispersant, thus obtaining a formulation containing 20% (w / w) of the AHAS-inhibiting herbicide.
[0155] C) Emulsifiable Concentrate (EC)
[0156] 15 par...
Embodiment 1
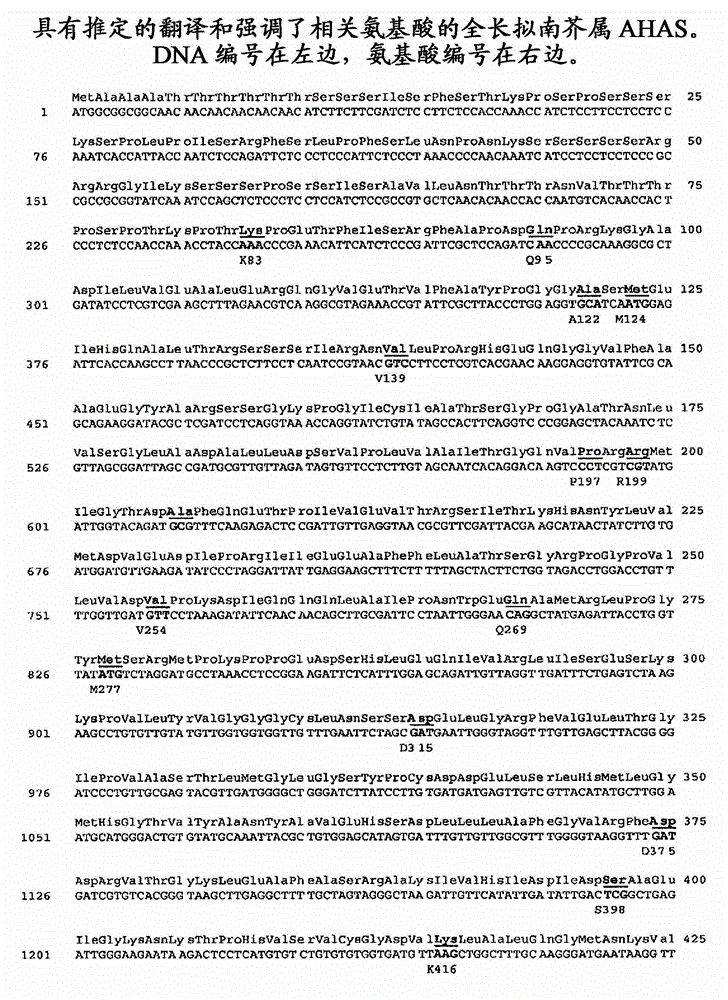
[0189] Vectors Containing Arabidopsis AHASL Mutant Genes
[0190] A complete XbaI fragment of Arabidopsis genomic DNA containing the complete AHAS large subunit gene with some additional DNA included at the 5' and an XbaI site at the 3' end. Bases 2484 to 4496 of SEQ ID NO: 34 include the coding sequence for the serine to threonine mutant allele at position 653 of the Arabidopsis AHAS large subunit gene, including the termination shown in SEQ ID NO: 30 a. The smaller of the serine to threonine mutant allele at position 653 of the Arabidopsis AHAS large subunit gene shown in SEQ ID NO:33 included in bases 2484 to 5717 of SEQ ID NO:34 The genomic fragment includes the coding sequence and the 3' end up to and including the 3' end Xbal site, where the first two bases of the NcoI site present at the start codon of AtAHASL were removed for clarity.
[0191] The DNA fragment SEQ ID NO:33 encoding the full-length Arabidopsis AHASL single mutant S653N and the 3' untranslated region ...
Embodiment 2
[0202] Embodiment 2: the carrier of maize AHASL mutant gene
[0203] The maize AHASL2 gene (SEQ ID NO:29) was cloned into the bacterial expression vector pTrcHis A (Invitrogen Corporation, Carlsbad, CA), fused with the vector tag and translation initiation site, starting from the 160th base of SEQ ID NO:29 . Using ZE1 as the base vector, mutagenesis and subcloning methods were used to generate vectors ZE2, ZE5, ZE6 and ZE7. A subcloning approach was used to generate ZE3 from ZE1, which is the maize AHASL2 gene fused to the vector tag and translation initiation site of pTrcHis A, starting at base 121 of SEQ ID NO:29. Since no functional differences between ZE1 or ZE3 were shown in E. coli, standard mutagenesis and subcloning methods were used to generate vectors ZE4 and ZE8 using ZE3 as the base vector.
[0204] A plant transformation vector with an expression cassette was prepared using standard methods and named ZP1 ( Figure 7 ), wherein the expression cassette comprises ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com