Method for discriminating two closely-related species of shellfish or identifying their hybrid generation
A kind of species and closely related technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of high cost, poor accuracy, and tediousness, and achieve the effect of intuitive results, simple methods, and wide application
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0048] Identification of Portuguese oyster C.angulata and Kumamoto oyster C.sikamea:
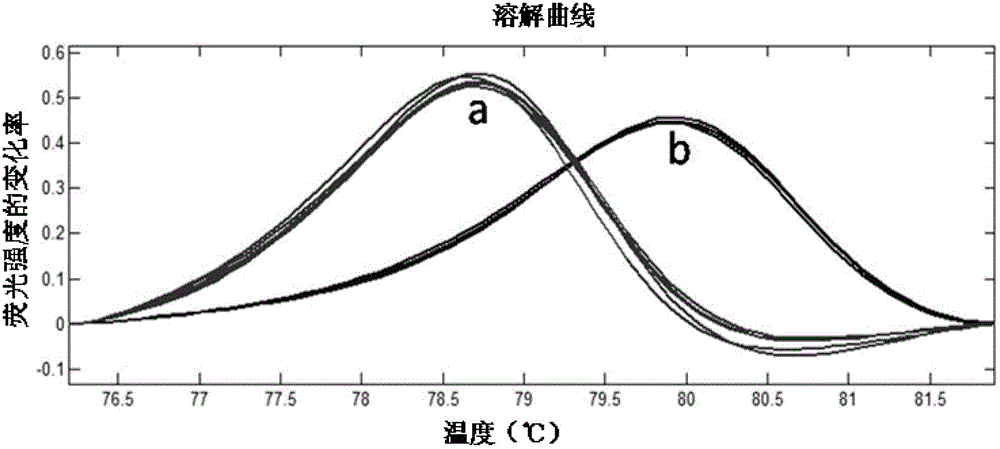
[0049] Portuguese oysters are also called mitral oysters in some areas of China. This oyster is only distributed along the coast south of the Yangtze River in China. It is the main economic species in southern China. Some areas, such as Putian, have also established the original breeding grounds and nature reserves of mitral oysters (actually the Portuguese oyster C. angulata). At the same time, the Kumamoto oyster C. sikamea is also the main species distributed in the Yangtze River estuary area and the coastal intertidal zone south of the Yangtze River, and it is often distributed in the same area as the Portuguese oyster. Some people also call this oyster the mitral oyster. Both types of oysters are small and easily confused. At present, the commonly used molecular biology identification method generally uses the mitochondrial COI gene sequence. In this case, six Portuguese oysters and ...
Embodiment 2
[0062] Identification of hybrids of two species:
[0063] Identification of hybrid progenies of Portuguese oyster C.angulata and Kumamoto oyster C.sikamea:
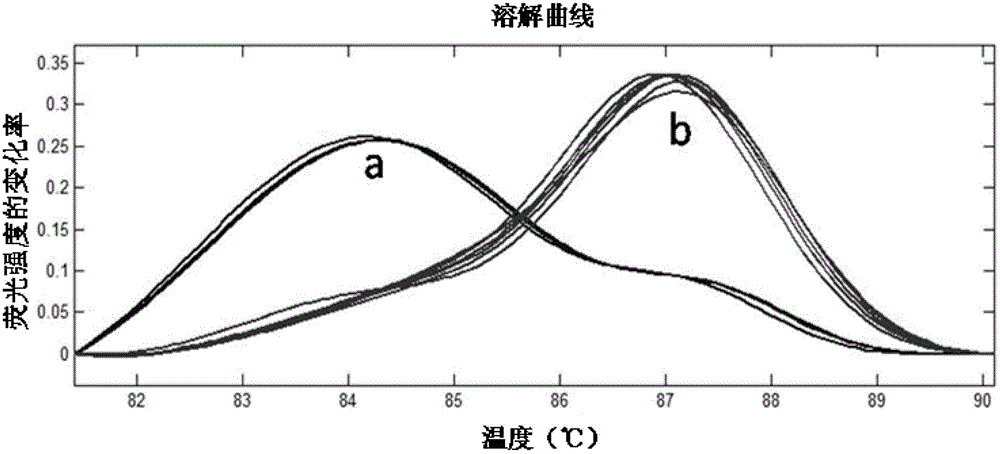
[0064] In many sea areas along the coast of China, oysters of different closely related species often live together in the same sea area. At the same time, due to the introduction of non-native oyster species from different regions, a large number of cultured oysters and local species live together at the same time. , resulting in the possibility of interbreeding. At present, there is no definite data showing the existence of hybrid populations, but in order to prevent the harm of local biological resources due to biological invasion, detection of interspecies hybridization is a top priority. Off the coast of Xiamen, China, Portuguese oysters and Kumamoto oysters often live in the same environment. In this case, artificial hybridization experiments were carried out on two species, and the hybrid offspring of the two spe...
Embodiment 3
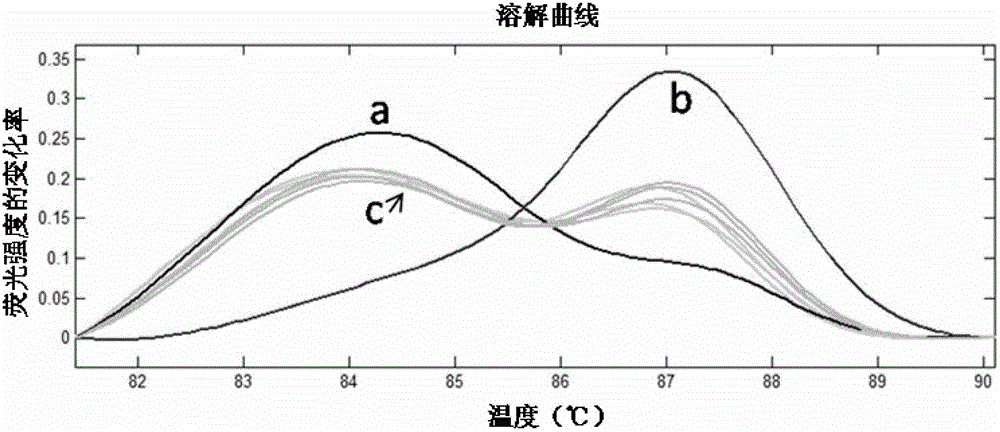
[0072] Use this method to identify closely related species: two species of oysters (Portuguese oyster C.angulata or Kumamoto oyster C.sikamea) were bought before the experiment. The oysters were named A oyster and B oyster, respectively, and they were Portuguese oyster C. angulata or Kumamoto oyster C. sikamea from the appearance.
[0073]a. Sequence acquisition: 14 and 12 ITS1 sequences of Portuguese oyster C. angulata and Kumamoto oyster C. sikamea were obtained from the NCBI database (http: / / www.ncbi.nlm.nih.gov / ). Alignment was performed by ClustalX software.
[0074] b. Primer design: Check the aligned sequences, and select InDel sites with stable differences between the two species based on the sequence characteristics of the ITS1 gene of the two species ( Figure 4 ), use Primer Premier5 software to design PCR primers on both sides of this site.
[0075] The primer sequences are:
[0076] ITS1-F1: 5'-AGCGCCTTGGGCCGTCGAA-3'
[0077] ITS1-R1: 5'-TGTACTTAAGGCGACGGAGC-3...
PUM
| Property | Measurement | Unit |
|---|---|---|
| diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com