Method for knocking out mir-505 from mammal cell line
A mammalian and cell line technology, applied in the field of knocking out mir-505 in mammalian cell lines, can solve problems such as unclear specific methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] (1) For the mir-505 gene, use the online sgRNA design software to design sgRNA
[0046]
[0047] sgRNA3: GC%: 52%;
[0048] sgRNA4: GC%: 43%
[0049] (2) In vitro synthesis of single-stranded nucleotide chains and formation of DNA double-stranded fragments
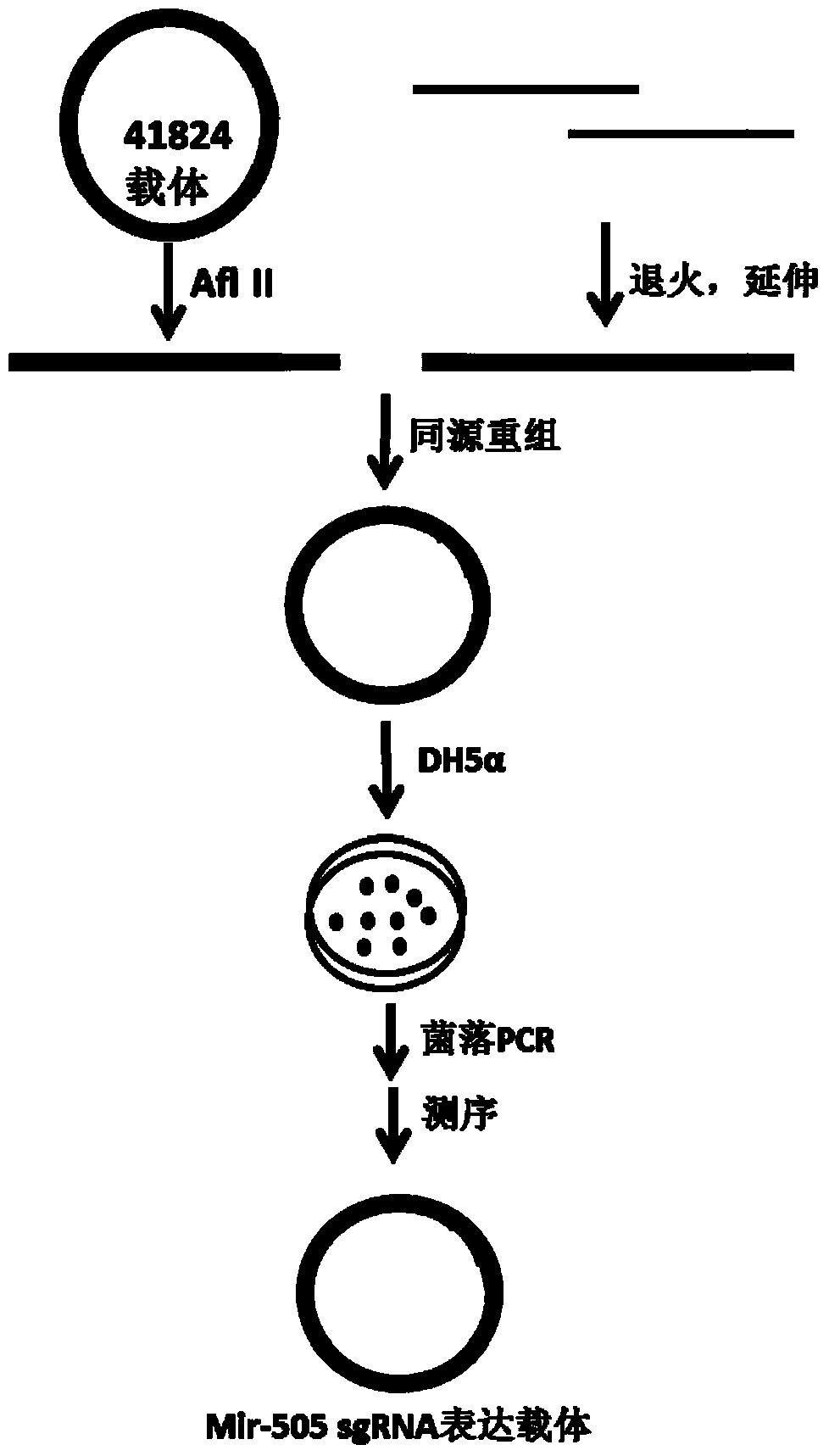
[0050] A. mir-505sgRNA3 insert in 41824
[0051] sgRNA3-F:5'-TTCTTGGCTTTATATATCTTGTGGAAAGGACGAAACACCGTAAATTGATGCACCCAGTG
[0052] sgRNA3-R:5'-ACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAACCACTGGGTGCATCAATTTAC
[0053] Using the PCR system, the two single-stranded nucleotides were annealed and extended into a double-stranded DNA fragment with a sequence length of 98bp, and identified by 2% agarose gel electrophoresis ( Figure 4 ). PCR system: add ddH2O 10ul, GT Buffer, 1.5ul, dNTP 1.5ul, positive and negative single-stranded (10P) 1.5ul, BSA 0.2ul, taq enzyme 1ul. PCR program: 95°C for 2min, 5 cycles of (95°C for 30sec, 56°C for 30sec, 72°C for 1min), 72°C for 10min.
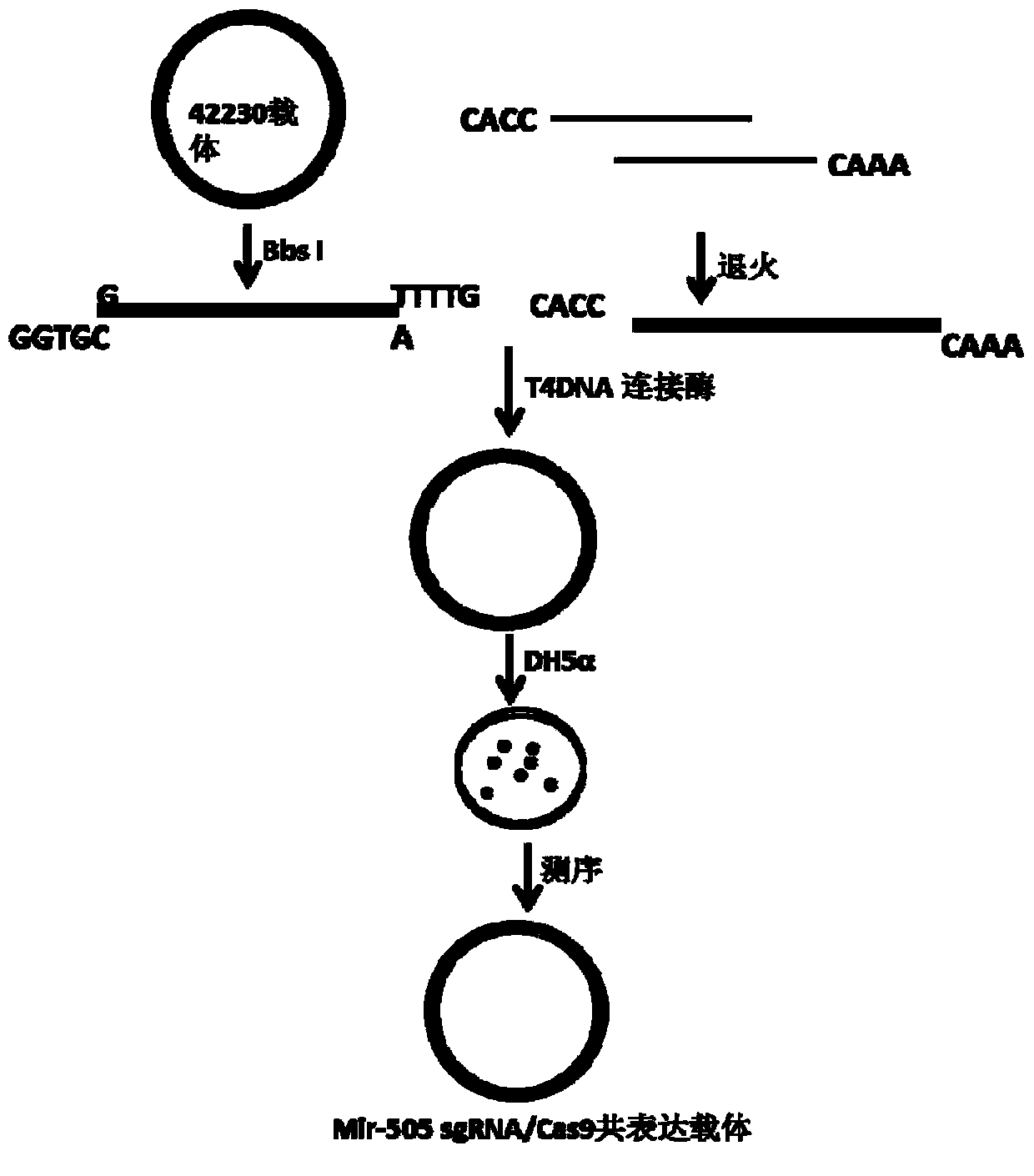
[0054] mir-505sgRNA3 and mir-505sgRNA4 inser...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com