3'-5' excision enzyme activity measurement method
An activity assay, exonuclease technology, applied in the field of biochemistry, can solve the problems of difficult screening, long cycle, difficult to achieve high-throughput, automation, etc., and achieve the effect of simple and fast operation steps
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
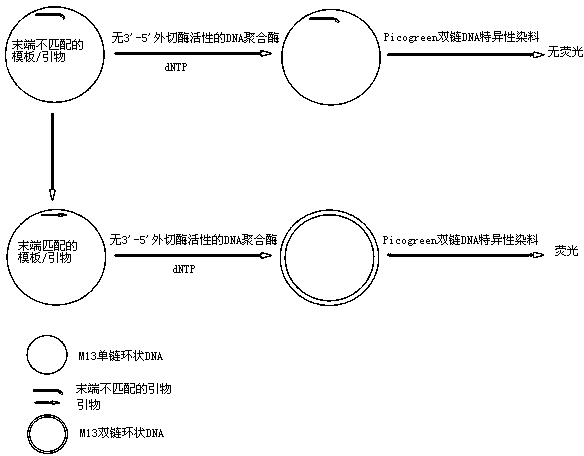
[0027] 1. Preparation of M13 template / primer complexes:
[0028] M13 ssDNA: M13mp18 ssDNA (NEB);
[0029] Primer: M13 primer 5'-aagccatccgcaaaaatgacctct-3';
[0030] M13 template / primer complex: M13mp18 single-stranded DNA was mixed with M13 primer at a molar ratio of 1:1, heated at 70°C for 5 minutes in annealing buffer (10 mM Tris-HCl pH 8.0, 50 mM NaCl), slowly Cool down to room temperature to anneal the template primers.
[0031] 2. Preparation of M13 template / mismatched primer complex:
[0032] M13 ssDNA: M13mp18 ssDNA (NEB);
[0033] Primer: M13 mismatch primer 5'-aagccatccgcaaaaatgacctct a -3'; (The underline indicates that the base does not match the template)
[0034] M13 template / mismatched primer complex: M13mp18 single-stranded DNA and M13 mismatched primer were mixed at a molar ratio of 1:1, heated at 70°C for 5 Minutes later, slowly cool down to room temperature to anneal the template primers.
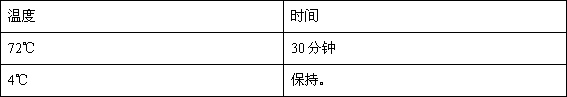
[0035] 3. 3'-5' exonuclease reaction:
[0036]
[0037] ...
Embodiment 2
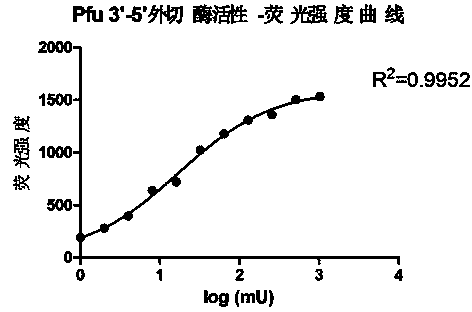
[0052] Example 2. Drawing of 3'-5' exonuclease activity-fluorescence intensity curve and EC50 calculation
[0053] Take the log value of the Pfu activity unit (mU) of 2#-12# as the abscissa, and the fluorescence intensity as the ordinate, and use GraphPad Prism5 to make a nonlinear regression curve. We found that these data points can fit a S-shaped curve very well, such as figure 2 shown.
[0054] By repeating the experiment several times and calculating the half-maximal effect concentration (EC50) of the curve, we found that the EC50 value remained stable across multiple experiments:
[0055]
[0056] Mean (mU) 17.75 Standard deviation SD (U) 0.47 Coefficient of Variation CV (%) 2.65
[0057] Therefore, this EC50 value can be used as the definition of Pfu 3'-5' exonuclease activity in this method. That is, in the reaction system of Example 1, the amount of enzyme that makes the fluorescence intensity reach half of the maximum value is define...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com