Nano-antibody aiming at SEA (Soluble Egg Antigen), and coding sequence and application thereof
A nanobody, sequence technology, applied in the field of biotechnology or biomedicine
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0020] This embodiment constructs the Schistosoma japonicum SEA-specific nanobody library, the steps are as follows:
[0021] (1) First, purify Schistosoma japonicum SEA, then mix 1 mg of SEA antigen with Freund's adjuvant in equal volume, and immunize an alpaca (Alpa-Vet, www.alpa-vet.be) once a week, 7 times in total , to stimulate B cells to express antigen-specific nanobodies;
[0022] (2) After the 7 times of immunization, extract 100ml alpaca peripheral blood lymphocytes and extract total RNA;
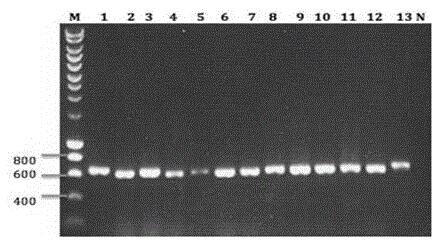
[0023] (3) Synthesize cDNA and amplify VHH by nested PCR;
[0024] (4) Digest 20ug of phage display vector and 10ul of VHH with restriction enzymes pstI and NotI and connect the two fragments;
[0025] (5) The ligation product was transformed into electroporation-competent cells TG1, and the SEA nanobody library was constructed and the storage capacity was determined. The storage capacity was 8.9×10 9 .
Embodiment 2
[0027] This embodiment screens SEA-specific Nanobodies, the steps are as follows:
[0028] (1) Dissolve in 100mM NaHCO 3 , 20ug SEA of pH 8.2 was coated on the NUNC microtiter plate, and placed overnight at 4°C;
[0029] (2) Add 100ul 3% milk the next day, and block at room temperature for 2 hours;
[0030] (3) After 2 hours, add 100ul 2×10 11 tfu contains the helper phage of the SEA nanobody library, and acts at room temperature for 1 hour;
[0031] (4) Wash 10 times with 0.05% PBS+Tween-20 for the first round of panning / 20-25 times for the second round / 20 times for the third round to remove non-specifically bound phages;
[0032] (5) Use 100mM TEA (triethylamine) to dissociate the phage that specifically binds to SEA, and infect Escherichia coli TG1 in the logarithmic growth phase, culture at 37°C for 1 hour, produce and purify the phage for the next round of screening, the same The screening process was repeated for 3-4 rounds, and the enrichment was obtained step by st...
Embodiment 3
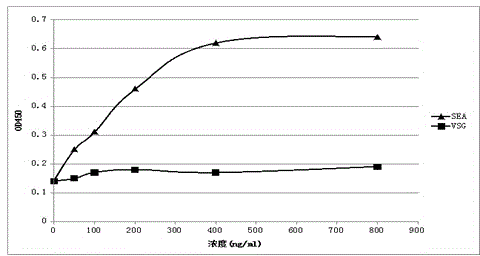
[0034] In this example, phage enzyme-linked immunosorbent assay (ELISA) was used to screen specific single positive clones, and the steps were as follows:
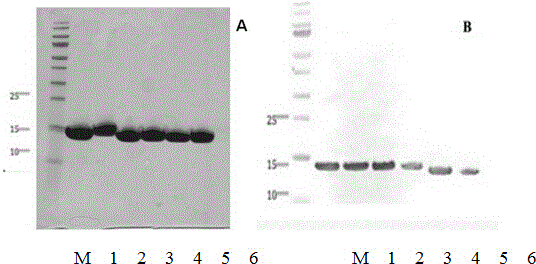
[0035] (1) From the cell culture dish containing phage after the above 3-4 rounds of screening, select 96 single colonies and inoculate them in TB medium containing 100ug / ml ampicillin, grow to the logarithmic phase, and add the final concentration 1 mM IPTG, culture overnight at 28°C.
[0036] (2) Use the infiltration method to obtain the crudely extracted antibody, transfer the antibody to an antigen-coated ELISA plate, and place it at room temperature for 1 hour;
[0037] (3) Wash off unbound antibody with PBST, add mouse anti-His antibody (mouse anti-HIS antibody, R&D system), and place at room temperature for 1 hour;
[0038] (4) Wash off the unbound antibody with PBST, and add anti-mouse alkaline phosphatase conjugate (goat anti-mouse AP labeled antibody, sigma).
[0039] (5) Wash off the unbound antibody with PBST...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com