Method for high-resolution genotyping of SSR (simple sequence repeats) molecular marker
A molecular marker and genotyping technology, applied in the field of molecular biology, can solve problems such as large labor force, low resolution, and increased cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
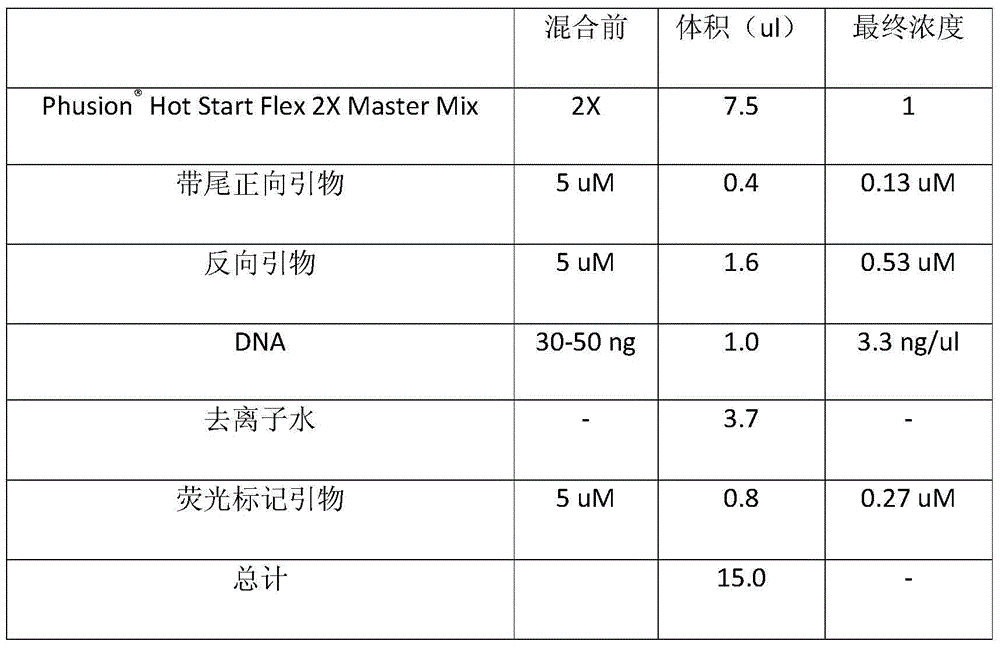
[0019] The following steps specifically describe the present invention from the design of SSR molecular markers, PCR formula, PCR reaction program, genotyping detection and data analysis based on sequencer:
[0020] (1). Design of three primers for SSR molecular markers
[0021] The present invention requires three primers: a universal fluorescent label primer, a tailed forward primer and a reverse primer. Universal fluorescent label primers are public primers, suitable for any PCR reaction in this method, and used in conjunction with tailed forward primers and reverse primers. The sequence of the fluorescent-labeled primer is fixed as CGTTGTAAAACGACGGCCAGT, and a fluorescent label needs to be added at the 5' end during primer synthesis. Optional fluorescent markers are any fluorescent reagents that can be recognized by ABI sequencers such as FAM, HEX, and TAMRA. Wherein, the choice of fluorescence cannot be the same as the fluorescence of the reference DNA molecule used for...
Embodiment 2
[0036] a. DNA extraction
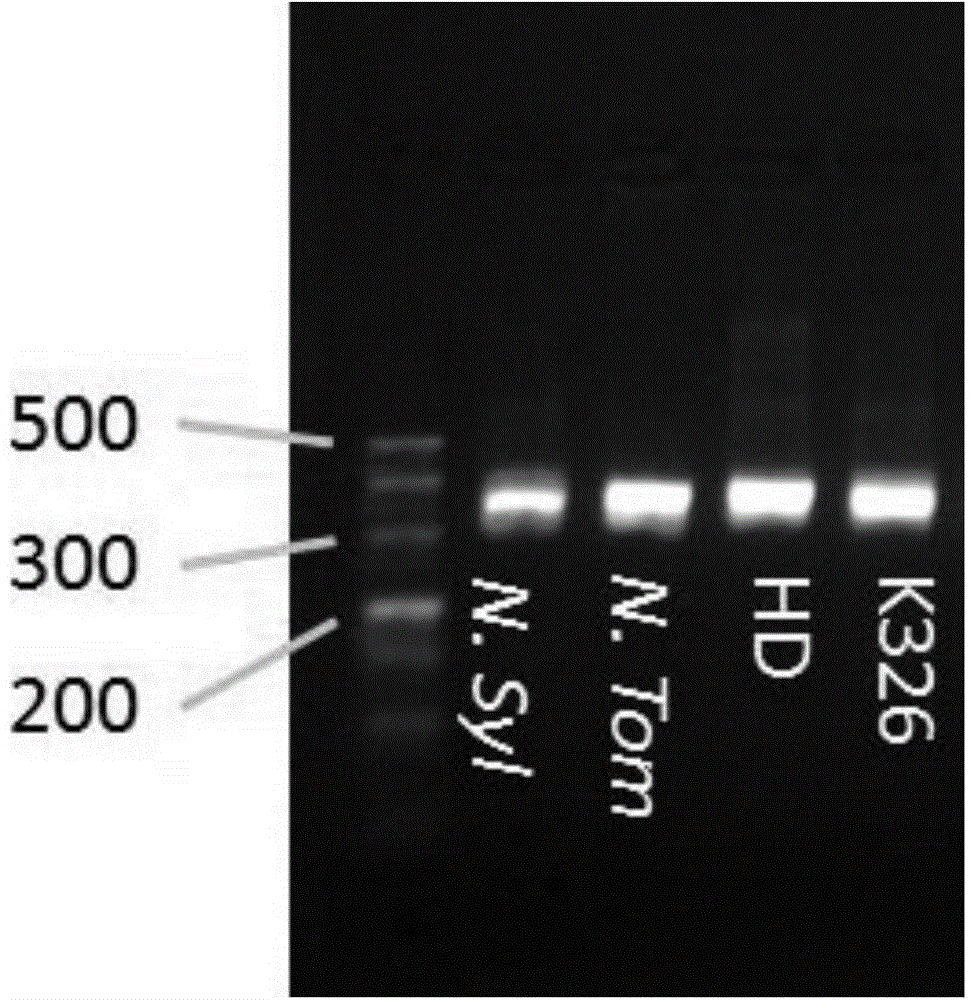
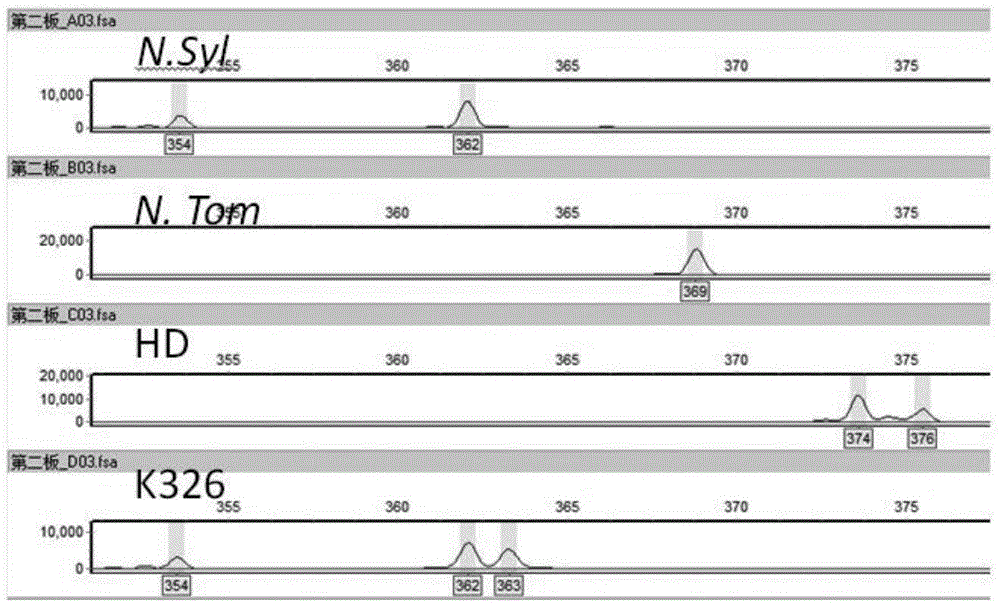
[0037] Five kinds of tobacco including tobacco forest, tobacco downy, HD tobacco and K326 tobacco were used as test samples. DNeasy Plant Mini Kit kit was used to extract the total DNA of each tobacco. The extraction method follows the steps provided in the kit, and finally dissolves the DNA to 50ul. Nanodrop detects DNA concentration. The concentration of DNA was diluted with redistilled water to a concentration of 30‐50ng / ul.
[0038] b. Design of three primers
[0039] According to the method described in Example 1, primers were designed using the online primer3 software and OligoAnalyzer 3.1. The sequences of the three designed primers are: 1. The tailed forward primer with the name xm8 is:
[0040] CGTTGTAAAACGACGGCCAGTCACTCGCAGTTTCTCTGCAC; 2. The reverse primer is:
[0041] ACCTCGAAAATCACCACCAG. 3. The sequence of the fluorescent labeling primer is: 5'FAM-
[0042] CGTTGTAAAACGACGGCCAGT. The designed primers were synthesized by Shangha...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com