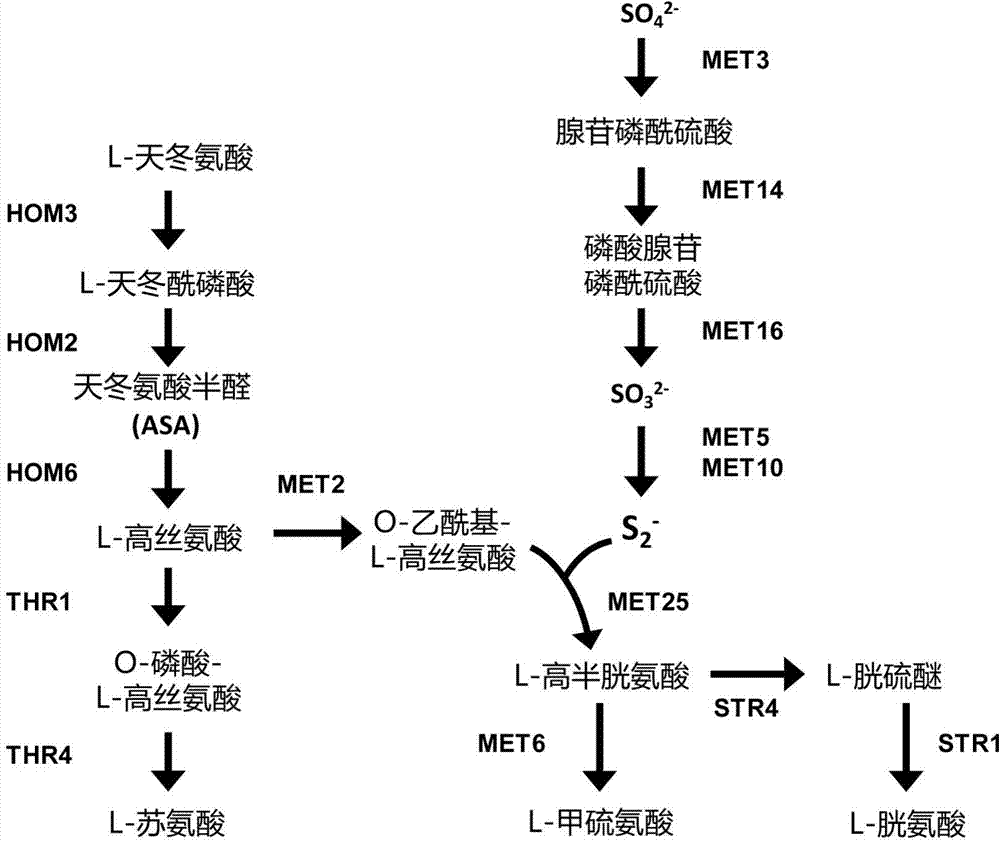
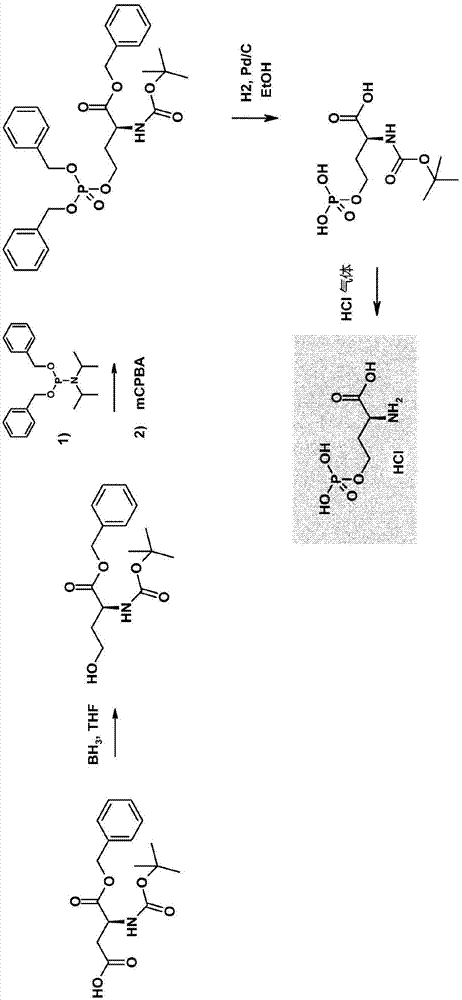
Process for producing L-methionine from O-phospho-L-homoserine and methanethiol employing a mutated cystathionine gamma-synthase
A technology of homoserine and methionine, applied in biochemical equipment and methods, DNA preparation, enzymes, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
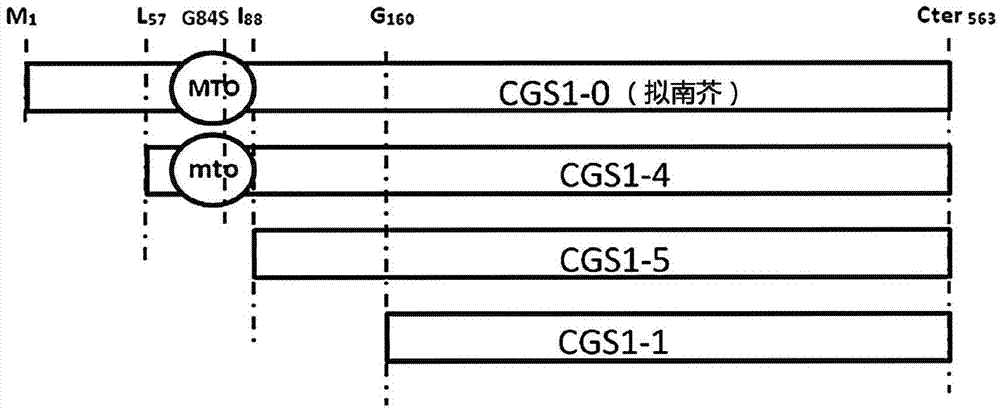
[0251] Example 1: Preparation of truncated forms of CGS1 used as starting material for the preparation of mutants
[0252]The CGS1-4 genes are synthetic genes. Synthesis was performed by Eurofins MWG Operon, (Ebersgerg) using the GENEius algorithm to optimize codon usage for improved expression in Saccharomyces cerevisiae. The S. cerevisiae codon usage table used was derived from the Kazusa Codon Usage Detabase (http: / / www.kazusa.or.jp / codon). Truncated forms of CGS1-5 and CGS1-1 have been obtained by using CGS1-4 as a substrate, using the forward oligonucleotide, GTACCGCTCGAG for CGS1-5 ATG GTTGCTGGTAAGTGGTCTAACAATC and GTACCGCTCGAG for CGS1-1 ATG TCTGTTCAATTGACCGATTCTAAG, prepared by PCR. For the CGS1-5 and CGS1-1 formats, use the equivalent reverse oligo AGTACGGGATCC TCA AATGGCTTCCA.
[0253] image 3 The truncated modified form of CGS1 is schematically represented in .
Embodiment 2
[0254] Example 2: Preparation of mutants of truncated forms of CGS1
[0255] A truncated version of the CGS1 gene was cloned into the pAL06 plasmid, a yeast replicating vector derived from the pRS316 vector (Sikorski RS & Hieter P., Genetics. 1989, 122: 19-27). The pAL06 plasmid allows expression of the cloned gene under the control of the strong yeast promoter TEF1. CGSA libraries were generated by hypermutation PCR at deoxynucleotide metaphosphate (dNTP) concentrations using the protocol described by Vartanian JP et al. (Nucleic Acids Res. 1996 24:2627-2631). [dTTP]>[dCTP] and [dGTP]>[dATP] biases are both used in mutation PCR. in 0.5mM MnCl 2 Use [dTTP] / [dCTP]=[dGTP] / [dATP]=1000 μM / 200 μM or [dTTP] / [dCTP]=[dGTP] / [dATP]=1000 μM / 150 μM.
Embodiment 3
[0256] Example 3: Screening of mutants that can prepare L-homoserine from O-phospho-L-homoserine and methylmercaptan
[0257] In yeast strains YA247-5A (MAT-α, ade2, his3, leu2, met2::loxP, met6::HIS3, trp1, ura3) or YA246-4A (MAT-a, ade2, his3, leu2, met2::loxP , met6::HIS3, thr4::loxP, trp1, ura3) in minimal medium supplemented with methionine but lacking uracil to transform the CGS1 library to select for plasmid markers. After 48 hours of growth, cells were harvested, washed and resuspended in minimal liquid medium containing methanethiol as the sulfur source. After 7 days, the culture was harvested and diluted with the same liquid medium to OD 600nm About 0.2. Three consecutive dilution cycles were performed.
[0258] The plasmids present in the resulting growing yeast cells were then extracted, expanded into E. coli and the resulting DNA used to retransform strain YA247-5A (MAT-α, ade2, his3, leu2, met2::loxP, met6::HIS3 , trp1, ura3) or YA246-4A (MAT-a, ade2, his3, l...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com