A dna molecular labeling method based on single primer amplification reaction
A DNA molecule, single-primer amplification technology, applied in the biological field, to achieve the effect of simple operation, abundant quantity and high stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
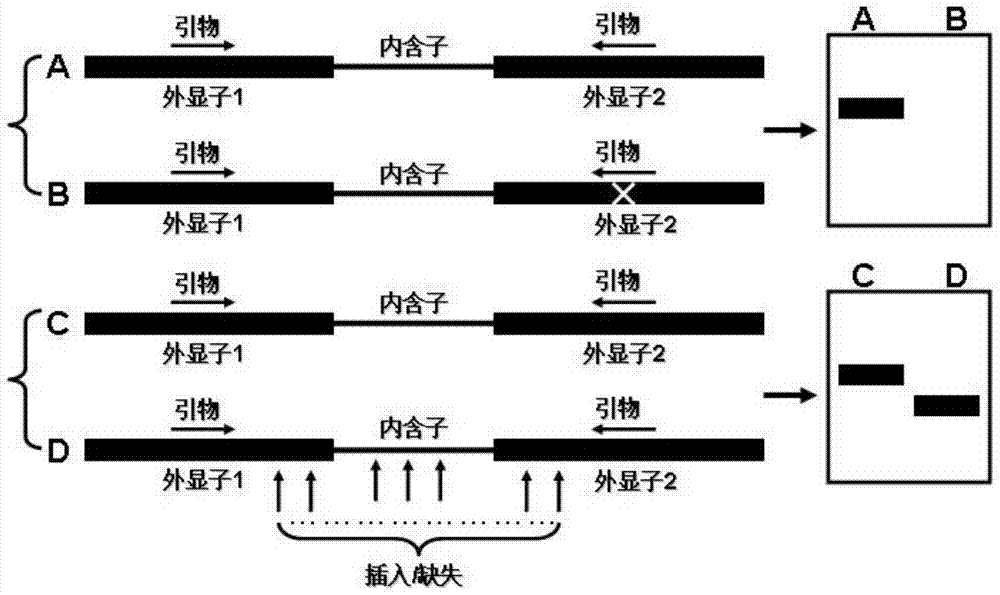
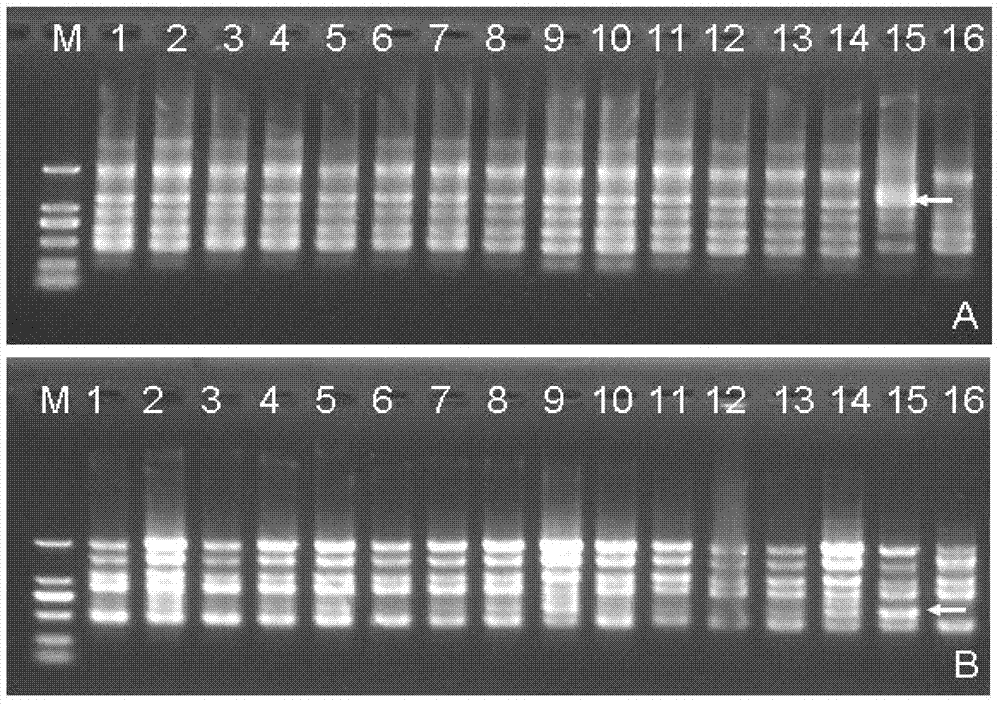
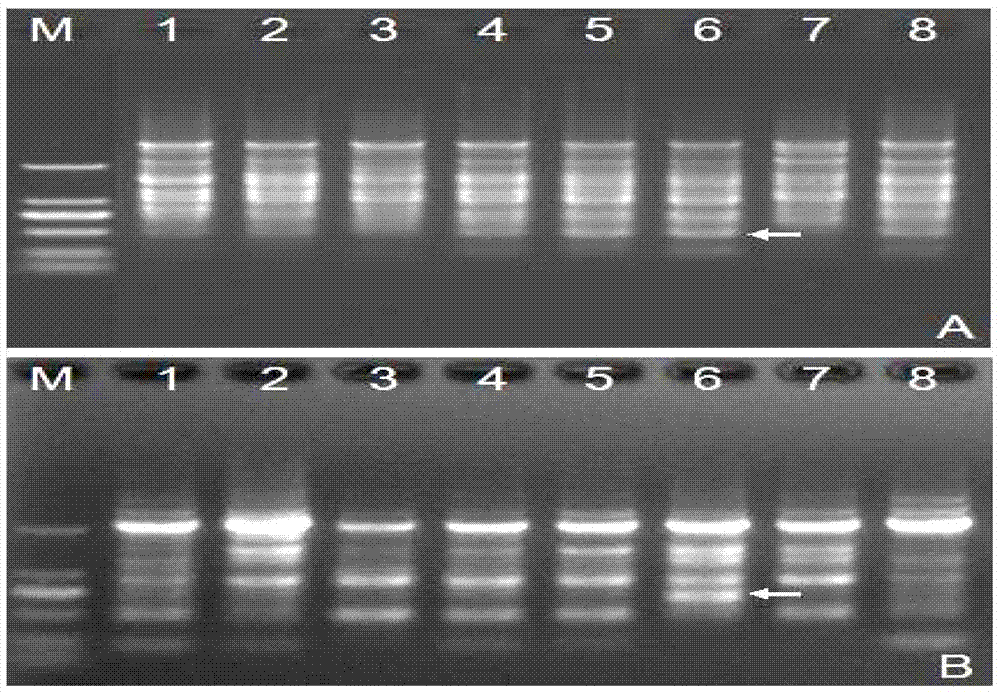
[0045] The schematic diagram of a DNA molecular labeling technique based on a single primer amplification reaction of the present invention is shown in figure 1 . The single primer used in this technology is designed based on the rich GC in the gene exon region of the eukaryotic genome. During PCR amplification, the single primer can simultaneously anchor and bind to the gene exon region in the genome The PCR product of the sequence between the gene exon binding sites in the eukaryotic genome is obtained to detect the polymorphism between the two binding sites in the gene exon region of the single primer. The polymorphism mainly comes from the following two aspects: one is the polymorphism caused by the point mutation at the primer binding site; the other is the length polymorphism caused by the sequence insertion, deletion and intron length difference between the primer binding sites Sex.
[0046] Hereinafter, the present invention will be further described in detail through th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com