SNP Molecular Markers of Pepper Ripening and Its Application
A molecular marker, pepper technology, applied in the field of pepper ripening SNP molecular markers, can solve the problems of large limitations in use, low phenotype selection accuracy, long genetic distance of target genes, etc., and achieves stable amplification and convenient detection. , the effect of speeding up the breeding process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0044] The development of embodiment 1 capsicum ripeness SNP molecular marker
[0045] 1. Population material construction and genetic analysis
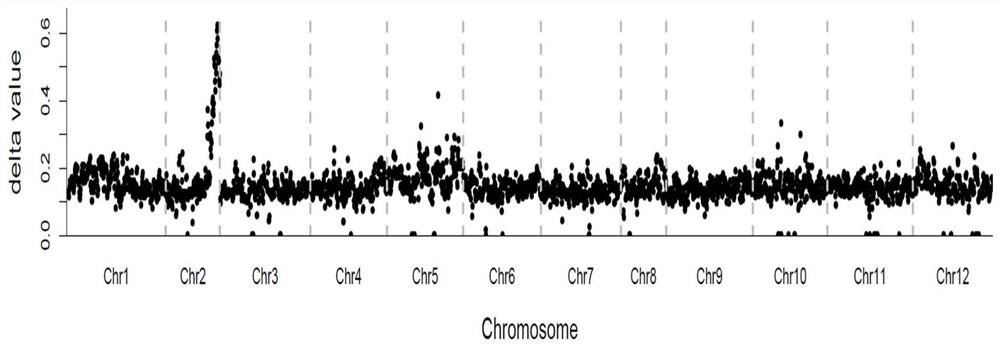
[0046] In the previous research process, the inventor found an early-maturing natural mutant strain in a population with normal maturity of pepper, and bred an early-maturing inbred line B with stable traits through multiple generations of selfing and directional selection. 9431 , depending on the planting environment, the first flowering node is 1-4 nodes. Take Early Pepper B 9431 As the female parent, late-maturing pepper (depending on the planting environment, the first flowering node is 13-18 nodes) A 145 F 1 Generation, F 1 Plants selfed to get F 2 group. Plant this F in spring 2017 2 There are 297 plants in the population, and the node position phenotype of flower initiation is measured by the number of true leaves (node positions) between the cotyledon and the first flower on the main stem. The statistical results sho...
Embodiment 2
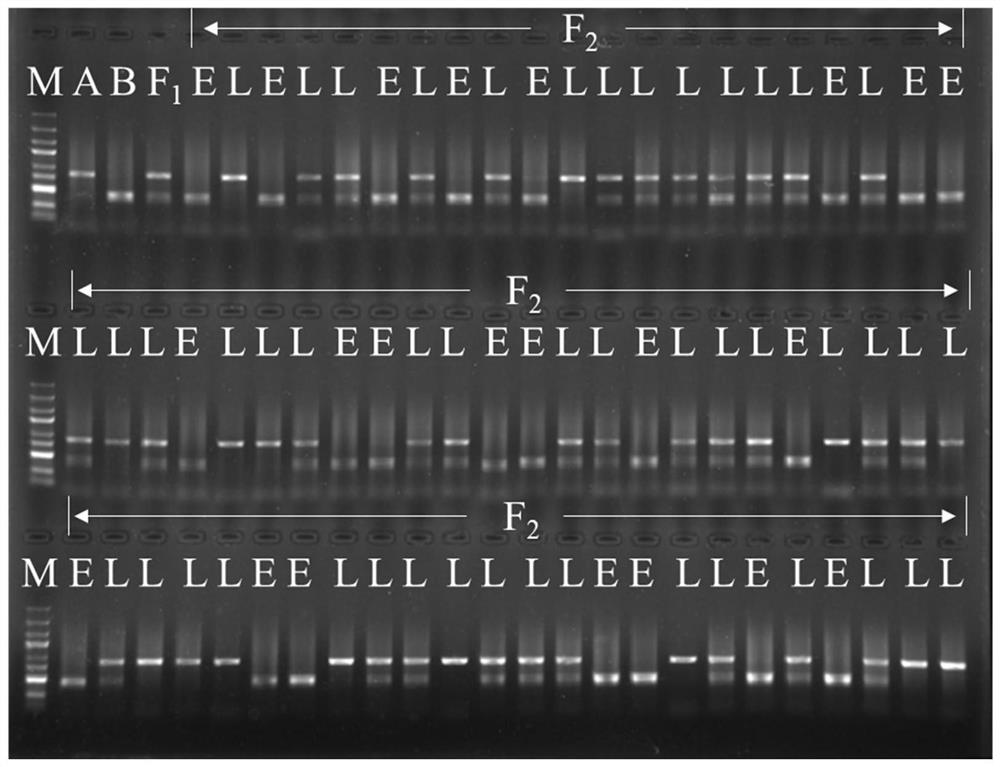
[0051] Example 2 Capsicum ripeness SNP molecular marker CSF2 in F 2 Application in the detection of colony plants
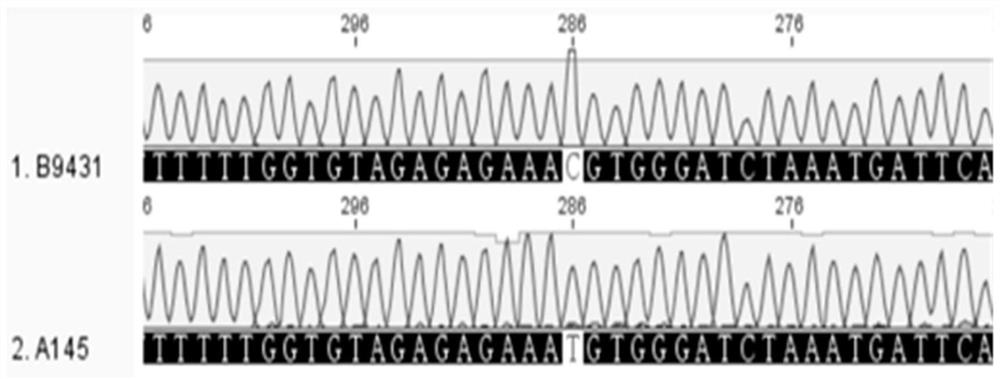
[0052] Extract parent B by CTAB method 9431 , parent A 145 , F 1 Plant (B 9431 ×A 145 ) and F 2 For plant DNA, the DNA concentration was detected with a Nanodrop spectrophotometer, and ddH was added 2 O Adjust all DNA concentrations to 100 ng / μL. Perform PCR according to the following conditions: Kangwei Century Biotechnology Co., Ltd. 2×Es Taq MasterMix (Dye) (CW0690) 5 μL, forward primer FFN1-5UF4 (10 μM) and reverse primer FFN1-e1R2 (10 μM) each 0.2 μL, 100ng 0.6 μL of DNA template per μL, add ddH 2 O to make up to 10 μL. PCR reaction program: pre-denaturation at 95°C for 2min; denaturation at 95°C for 30s, annealing at 55°C for 30s, extension at 72°C for 40s, 38 cycles; extension at 72°C for 10min. The PCR reaction product was digested with restriction endonuclease TaiI: 0.65 μL of 10×Buffer R, 3.5 μL of PCR product, 0.1 μL of TaiI restriction endon...
Embodiment 3
[0053] Example 3 Application of Pepper Ripeness SNP Molecular Marker CSF2 in Identifying Pepper Ripeness Traits
[0054] 92 pepper (Capsicum annuum) germplasm materials (Table 1) with a variation range of 5-23 nodes of the first flowering node were selected, and their DNA was extracted by CTAB method, and the extra-early-maturing pepper B 9431 , Late Pepper A 145 and its F 1 Plant (B 9431 ×A 145 ) DNA was used as a control, and PCR was carried out according to the following conditions: the total reaction volume was 10 μL, including 5 μL of Kangwei Century Biotechnology Co., Ltd. 2×Es TaqMasterMix (Dye) (CW0690), forward primer FFN1-5UF4 (10 μM) and reverse primer 0.2 μL each of FFN1-e1R2 (10 μM), 0.6 μL of 30-100 ng / μL DNA template, add ddH 2 O to make up to 10 μL. PCR reaction program: pre-denaturation at 95°C for 2min; denaturation at 95°C for 30s, annealing at 55°C for 30s, extension at 72°C for 40s, 38 cycles; extension at 72°C for 10min. The PCR reaction product was...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com